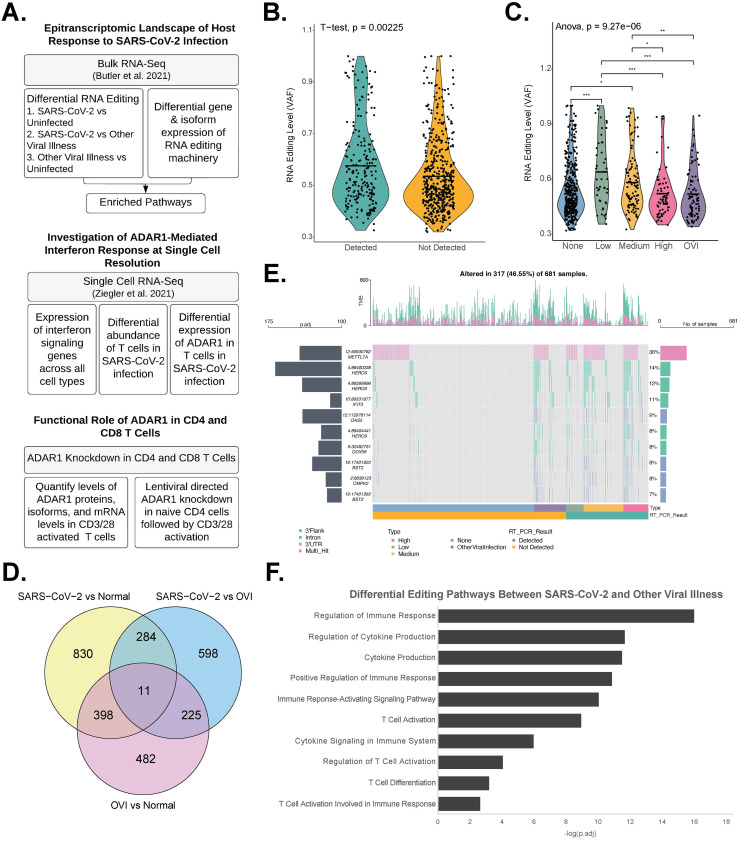
Fig 1. Differential editing and enriched pathways in SARS-CoV-2 nasopharyngeal swab samples.
a. Overview of workflow. b. Overall RNA editing levels are significantly higher in SARS-CoV-2 positive patients than negative patients (p = 0.00225). c. RNA editing levels increase with decreasing viral load (p = 9.27E-6). * p < 0.05; ** p < 0.01; *** p < 0.001. d. Venn diagram of differential RNA editing sites found in three comparisons: SARS-CoV-2 positive vs normal: 1,523; other viral illness vs normal: 1,116; SARS-CoV-2 positive vs other viral illness: 1,118. e. Differential editing sites between COVID-19 positive and negative samples are found in introns, the 3’ UTR, and the 3’ Flank. f. Pathway analysis of differentially edited genes between SARS-CoV-2 and other viral illness samples (adj.p < 0.05).