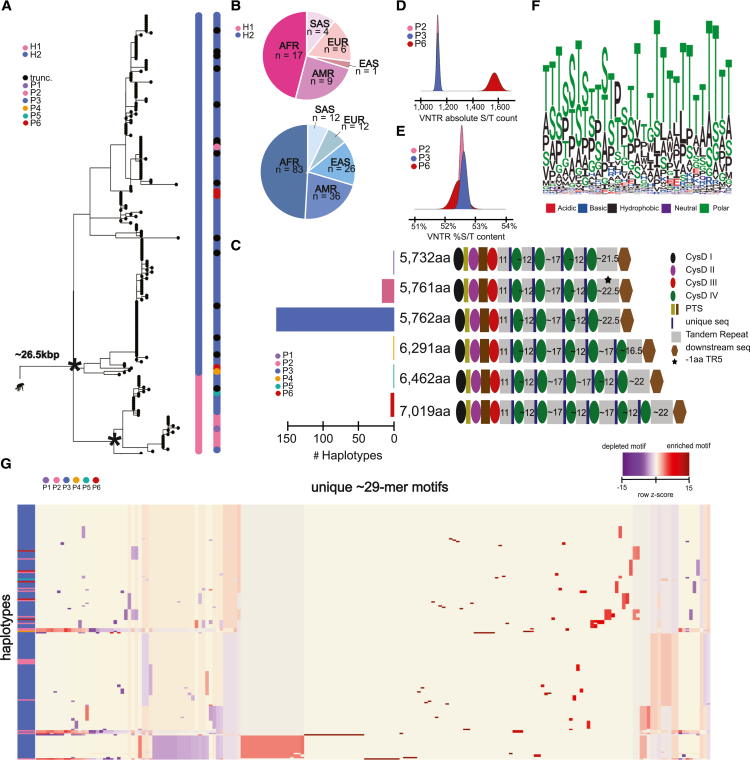
Figure 2.
The genetic architecture of MUC5B in 206 human haplotypes
(A) Recombination-aware phylogenetic analysis of ∼26.5 kbp neutral sequence (introns 16–48) from 206 human haplotypes of MUC5B with two chimpanzee haplotypes as outgroup. (∗) = central node with 100% bootstrap support. H1 and H2 correspond to two major haplogroups; P1–P6 correspond to protein groups (consistent with C); trunc. corresponds to haplotypes with truncated protein predictions.
(B) Frequency of population-specific haplotypes found in the two common phylogenetic haplogroups of MUC5B.
(C) Protein predictions for 206 human haplotypes of MUC5B. Diagrams represent protein domains with the large central exon of MUC5B, modeled after those in Ridley et al.53 Colors correspond to protein groups visualized in (A). CysD corresponds to cys domains and PTS corresponds to proline-, serine-, and threonine-rich domains.
(D) Distributions of absolute serine and threonine (S/T) count across VNTR domains for the three most common protein groups of MUC5B.
(E) Distributions of percent S/T content within VNTR domains for the three most common protein groups of MUC5B.
(F.) Logo plot of the complete 29-mer amino acid motif variants used in MUC5B VNTR domains across 206 human haplotypes. Colors correspond to biochemical groupings of amino acids.
(G) Heatmap of 190–29-mer motif utilization across protein variants of human MUC5B, colored vertically by protein group identities. Heatmap constructed through normalization for total VNTR sequence length, normalization within each motif (columns), and hierarchical clustering of haplotypes (rows) and motifs (columns). See Figure S4 for an extended version that includes the matched motifs (columns).