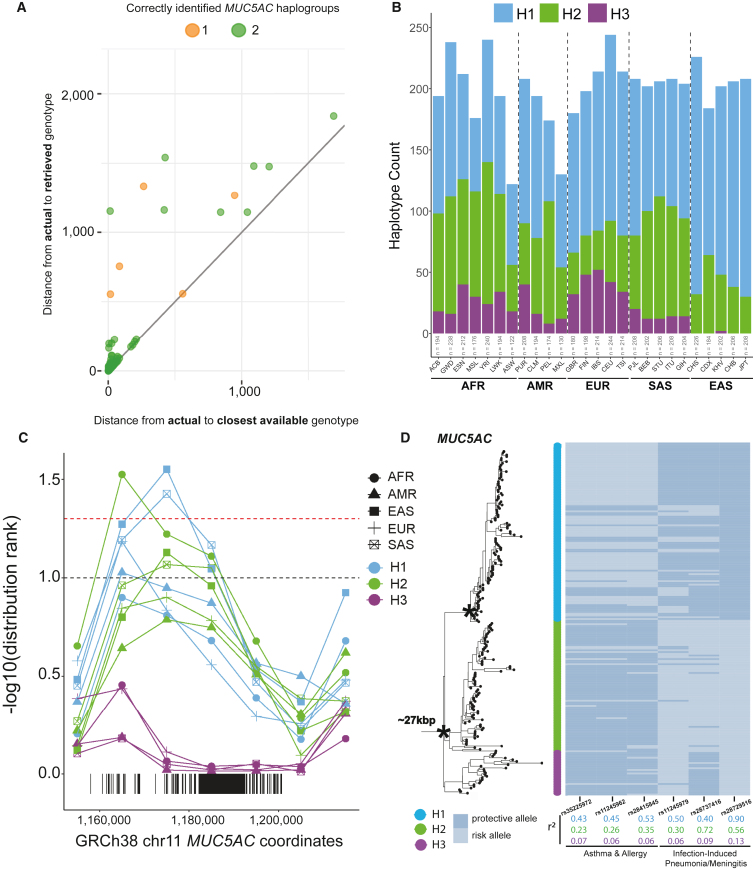
Figure 5.
Genotyping of MUC5AC haplogroups with Locityper for population distributions and signatures of positive selection
(A) Locityper leave-one-out results comparing edit distances between actual and retrieved genotype (predicted from Locityper) versus edit distances between actual and closest possible genotype (best possible reference genotype from a multiple sequence alignment with true genotype) for MUC5AC. Dot color based on the number of haplotypes in diploid sample sets that were correctly genotyped.
(B) MUC5AC haplogroup frequencies across super populations and populations in the 1KG dataset from Locityper predictions.
(C) Distribution ranks of negative Tajima’s D values across 10 kbp bins in the MUC5AC locus for genotyped haplogroups in each of the 1KG super populations. The dashed black line corresponds to the 10% distribution rank and the dashed red line corresponds to the 5% distribution rank. The three values above the dashed red line pass permutation testing and multiple testing correction.
(D) Six GWAS risk and protective alleles mapped to the MUC5AC phylogeny. SNPs grouped based on disease association and squared correlations color coded based on haplogroup partitioning.