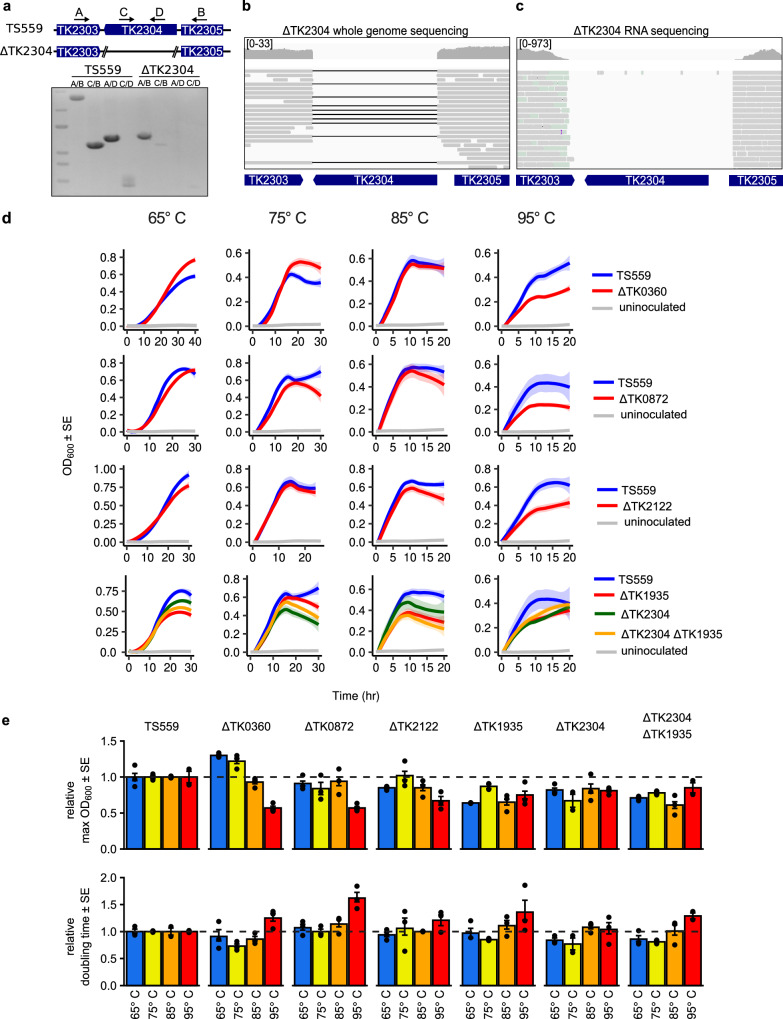
Fig. 2. The m5C epitranscriptome supports hyperthermophilic growth.
a The deletion of gene TK2304 was initially confirmed by PCR using 4 sets of primers. External primers (A/B) amplify across the deleted locus and result in a full length amplicon in parent strain TS559 and an amplicon reduced by the size of gene TK2304 in the ΔTK2304 strain. Internal primers (C/D) or a combination of internal and external primers (A/D, C/B) result in amplification in strain TS559 but not ΔTK2304. An uncropped gel image is provided in Source Data. Final confirmation was performed using (b) Minion whole genome sequencing (DNA-seq) and (c) Illumina RNA bisulfite sequencing (RNA-seq). Visual inspection of DNA and RNA sequences aligned to the T. kodakarensis reference genome using Integrative Genomics Viewer show the absence of reads from the deleted loci. In the DNA-seq windows, black lines connect contiguous reads and indicate a gapped alignment. The coverage range for each window is listed in brackets. d Head-to-head growth competitions were performed at 65°, 75°, 85°, and 95 °C for each strain deleted for an R5CMT (ΔTK0360, ΔTK0872, ΔTK2122, ΔTK1935, ΔTK2304), the double deletion (ΔTK1935 ΔTK2304), and parent strain TS559. e The maximum optical density (Max OD600) and the rate of growth (doubling time) for each culture is illustrated relative to parent strain TS559. ± 1 standard error (SE) is represented by error bars for n = 3 biological replicates.