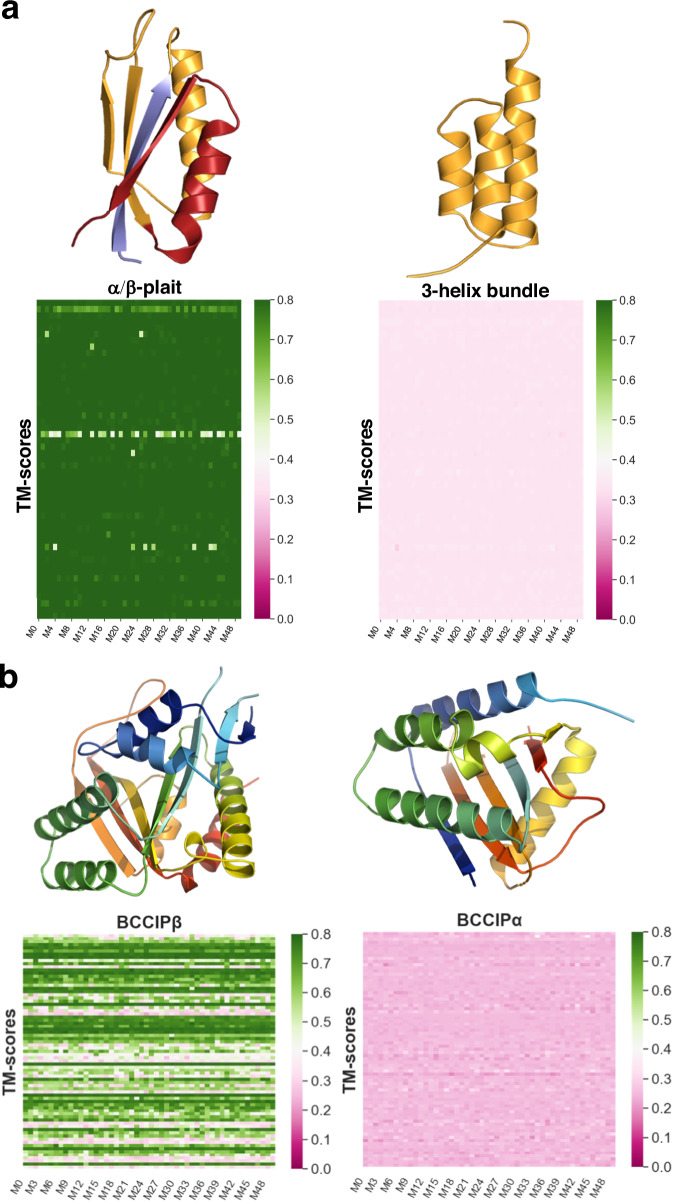
Fig. 3. AF2 fails to predict fold switching of two protein structures outside of its training set.
a Sa1 is a designed protein that switches reversibly between α/β-plait (PDBID:8e6y, Fold1) and 3α helix (PDBID: 2fs1, Fold2) folds triggered by temperature changes. Cartoon representations of Fold1 are colored blue for N-terminal residues (1 to 10), orange for the fold-switching residues (11 to 66 aligning with the amino acid sequence in Fold2, also in orange) and C-terminal residues (67 to 95) are red. Heatmaps of 50 predictions (M0 to M49) for each of 51 sequence clusters showing the similarity (TM-scores) to Fold1(left panel) and Fold2 (right) are presented below the cartoon representations of the two states. AF-cluster consistently predicts Fold1 but misses Fold2. b BCCIPβ and BCCIPα are human protein isoforms with 80% sequence identity that adopt distinct folds. (13 Å RMSD). AF-cluster was run on BCCIPα’s sequence. In the right panel, a cartoon representation of BCCIPα (colored blue to red from N-terminus to C-terminus) is shown with the heatmap of TM-scores of 50 predictions (model numbers M0 to M49) for each of 75 sequence clusters compared to the fold adopted by the α isoform (PDBID:8exf, chain B). In the left panel, the BCCIPβ experimental structure (PDBID:7kys) is shown with the heatmap of TM-scores compared to the fold adopted by the β isoform. AF-cluster frequently predicts the structure of the β-isoform but misses the experimentally consistent α-isoform structure.