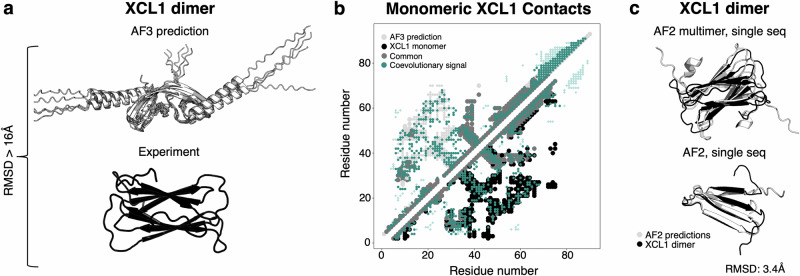
Fig. 5. AF predicts the dimeric form of XCL1 through structure memorization rather than coevolutionary inference.
a Though AF3 was given appropriate stoichiometry and environmental conditions to predict the lymphotactin dimer, its prediction did not match experiment. b The coevolutionary patterns of AF3’s predicted XCL1 dimer match those of its monomeric conformation almost exactly. Contact maps generated from the AF3 prediction and experimentally determined monomeric conformation (2hdm). Upper diagonal corresponds to contacts unique to the AF3 prediction (light gray smaller dots correspond to intermolecular contacts; larger to intramolecular), lower diagonal corresponds to contacts unique to the experimentally determined monomeric conformation (black); common contacts medium gray; coevolutionary information inferred from MSA using ACE, (teal). c Both AF2 multimer and AF2 predict the correct XCL1 dimer structure from single sequences and 0 recycles, suggesting that they memorized its structure during training.