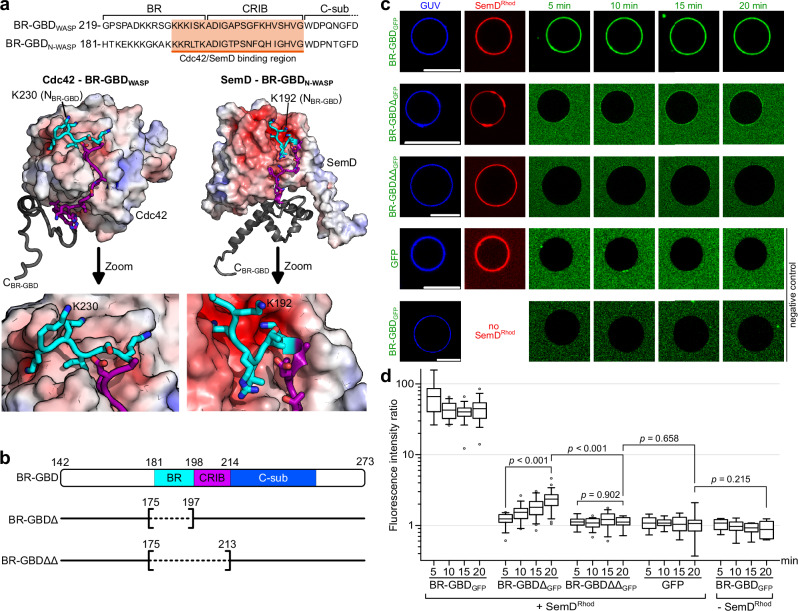
Fig. 3. SemD recruits the BR-GBD region of N-WASP to membrane vesicles.
a Amino acid sequence of WASP219-259 (human) and N-WASP181-221 (Rattus norvegicus) (of which the latter is identical to N-WASP184-224 from human) (top). The orange box shows the sequence involved in binding to Cdc42GTP and SemD, respectively. The lower panel shows the structures of Cdc42GTP in complex with BR-GBDWASP (PDB: 1CEE37) and SemDΔAPH bound to BR-GBDN-WASP. Cdc42GTP binds to the positively charged C-terminal KKK230-232 motif of the BR from WASP and embeds the CRIB domain in the less charged binding groove. The same binding mechanism is observed for SemD, in which a negatively charged patch engages with the positively charged KKR192-194 motif within the N-WASP BR domain and inserts the subsequent N-WASP CRIB domain into the SemD binding groove. b Schematic representation of BR-GBD and the deletion variants BR-GBDΔ (lacking the BR175-197 domain) and BR-GBDΔΔ (lacking the BR175-197 and the CRIB198-213 domains). Dashed lines in square brackets mark the deleted protein regions. The first and last deleted amino acids are indicated. c Confocal images of PS-containing GUVs with rhodamine-labelled SemD (SemDRhod) and BR-GBD variants fused to GFP (BR-GBDGFP). (scale bars 10 µm). d Quantification of bound protein to the GUV membrane based on the ratio of the maximal fluorescence at the perimeter of the GUV to the average background fluorescence outside the GUV. For each variant and time point, the fluorescence intensity ratio was calculated for up to 22 independent GUVs. The data are represented as boxplots with whiskers. The boxes are limited by the 25th and 75th percentile, including 50% of the data. The centre line shows the mean score. Whiskers denote 5–95% of all data and outliers are shown as grey dots. For comparing two groups, an unpaired, two-sided Student’s t test was used. The data are representative for two independent data sets, both yielding similar results. Source data of both data sets are provided as Source Data file.