Highlights
-
•
An odorant receptor from the American palm weevil detects aggregation pheromones of other species.
-
•
The response spectrum of this odorant receptor is highly conserved in the American and the Asian palm weevils.
-
•
In both species, this receptor is broadly tuned and is also activated by host plant volatiles.
Keywords: Aggregation pheromone, Odorant receptors, Palm tree volatiles, 3D structure modelling
Abstract
The evolution of chemosensory receptors is key for the adaptation of animals to their environment. Recent knowledge acquired on the tri-dimensional structure of insect odorant receptors makes it possible to study the link between modifications in the receptor structure and evolution of response spectra in more depth. We investigated this question in palm weevils, several species of which are well-known invasive pests of ornamental or cultivated palm trees worldwide. These insects use aggregation pheromones to gather on their host plants for feeding and reproduction. An odorant receptor detecting the aggregation pheromone components was characterised in the Asian palm weevil Rhynchophorus ferrugineus. This study compared the response spectra of this receptor, RferOR1, and its ortholog in the American palm weevil R. palmarum, RpalOR1. Sequences of these two receptors exhibit more than 70 amino acid differences, but modelling of their 3D structures revealed that their putative binding pockets differ by only three amino acids, suggesting possible tuning conservation. Further functional characterization of RpalOR1 confirmed this hypothesis, as RpalOR1 and RferOR1 exhibited highly similar responses to coleopteran aggregation pheromones and chemically related molecules. Notably, we showed that R. ferrugineus pheromone compounds strongly activated RpalOR1, but we did not evidence any response to the R. palmarum pheromone compound rhynchophorol. Moreover, we discovered that several host plant volatiles also activated both pheromone receptors, although with lower sensitivity. This study not only reveals evolutionary conservation of odorant receptor tuning across the two palm weevil species, but also questions the specificity of pheromone detection usually observed in insects.
1. Introduction
The evolution of chemical senses is a major element in the adaptation of animal behaviour to their environment. Taste and olfaction can evolve rapidly, for example in relation to changes in feeding habits or in abilities to communicate with conspecifics. This evolution relies on modifications in neural circuits (Zhao and McBride, 2020) and gene families that govern detection capacities in the peripheral nervous system (Baldwin and Ko, 2020; Robertson, 2019; Vizueta et al., 2020). In insects, the main gene family that establishes the olfactory detection capacities is the odorant receptor (OR) family, which does not exist in other animal taxa (Suh et al., 2014). ORs are 7-transmembrane receptors expressed in olfactory sensory neurons (OSNs) found mainly in antennae, where they allow for signal transduction. Insect ORs assemble in heteromeric odorant-gated ion channels composed of a ligand-selective variable subunit and a unique co-receptor subunit named Orco (Benton et al., 2006).
Whereas Orco is highly conserved across insects, insect OR gene repertoires can diversify rapidly following a birth-and-death model of evolution. On a broad timescale, this translates into highly divergent OR repertoires between insects from different taxa, with gene numbers ranging from 10 to more than 300 and no detectable gene homologies (Robertson, 2019). Across shorter time scales, it has been shown that paralogs resulting from OR gene duplications can diverge fast, leading to functional divergence (Hou et al., 2021). On the other hand, orthologs can exhibit a highly conserved response spectrum despite a sequence divergence above 30 % (Guo et al., 2021; Roberts et al., 2022), but only one or two mutations can be enough to shift the response spectra between pairs of orthologs, as demonstrated for moth pheromone receptors (Cao et al., 2021; Leary et al., 2012; Yang et al., 2017). The lack of a resolved tri-dimensional (3D) structure for an insect OR has hindered studies on the structural bases of OR evolution for a long time. The recent elucidation of the 3D structures of one Orco (Butterwick et al., 2018) and one OR subunit (del Mármol et al., 2021), as well as the advent of efficient in silico modelling tools such as AlphaFold2 (Jumper et al., 2021), now make it possible to better predict and interpret the functional consequences of amino acid changes within ORs, opening up new perspectives for understanding their evolution (Cao et al., 2023; Li et al., 2023).
With more than 80,000 species, Curculionidae beetles (gathering weevils and bark beetles) display an unparalleled diversity and constitute the largest family of animals on Earth. These phytophagous insects are adapted to many host plants, and some are serious pests of crops or forests. They make use of aggregation pheromones, which are attractive semiochemicals detected at long distances that allow adults to cluster at their feeding and breeding sites. In the context of reducing the use of pesticides for crop protection, aggregation pheromones and host plant volatiles have been identified in several bark beetles (subfamily Scolytinae) and palm weevils (subfamily Dryophthorinae) and further used as trap baits in integrated pest management (Bandeira et al., 2021; Giblin-Davis et al., 1996; Oehlschlager, 2016). Consequently, they have become models for the study of olfaction. Molecular mechanisms of olfaction have remained unknown for long in these insects but the recent functional characterization of ORs in the European spruce bark beetle Ips typographus (Yuvaraj et al., 2021) and in the Asian palm weevil Rhynchophorus ferrugineus (Antony et al., 2021, Antony et al., 2024; Ji et al., 2021) have paved the way for OR evolutionary studies (Hou et al., 2021; Roberts et al., 2022). Notably, the recent sequencing of the antennal transcriptome of the American palm weevil R. palmarum (Gonzalez et al., 2021) enables comparison of receptors from these two closely related weevils.
R. ferrugineus originates from South East Asia and has spread through the Middle East, Africa, and the Mediterranean basin in the late 20th century (Hoddle et al., 2024). It is the most invasive and destructive pest of palm trees worldwide. The aggregation pheromone released by males is a mix of (4S,5S)−4-methylnonan-5-ol and (S)−4-methylnonan-5-one, known as ferrugineol and ferrugineone, respectively (Hallett et al., 1993; Perez et al., 1996). R. palmarum is an important pest of coconut and oil palm trees in Central and South America (Hoddle et al., 2024). It causes damage because of larval feeding into the stems but also because it is the primary vector of the nematode Bursencephalus cocophilus, the agent of the red ring disease that strongly affects the palm trees (Gerber and Giblin-Davis, 1990). R. palmarum male adults produce an aggregation pheromone made of (4S,2E)−6-methylhept-2-en-4-ol, which is referred to as rhynchophorol (Oehlschlager et al., 1992; Rochat et al., 1991). In both species, synthetic mixtures of (S) and (R)-enantiomers of the aggregation pheromone components are commonly used in pheromone-baited traps in combination with plant tissues such as pieces of decaying palm stems, coconut, sugarcane or pineapple, which increase attraction due to a synergistic effect between the pheromone and host plant volatiles (Abbas et al., 2006; Guarino et al., 2011; Jaffé et al., 1993; Oehlschlager et al., 1993; Rochat et al., 2000).
We recently identified an aggregation pheromone receptor in R. ferrugineus, named RferOR1. This OR was highly sensitive to the two components of the pheromone but was also activated by pheromone compounds from other beetles (Antony et al., 2021). Sequencing of the antennal transcriptome of R. palmarum revealed a RferOR1 ortholog named RpalOR1 (Gonzalez et al., 2021). RpalOR1 was among the most highly expressed ORs in both males and females, suggesting an important role in the ecology of R. palmarum. As RpalOR1 and RferOR1 exhibited ∼80 % amino acid sequence identity, we wondered whether their response spectra would be conserved between these two Rhynchophorus species. Here, we used a combination of 3D-modelling and functional analyses through heterologous expression in Drosophila OSNs to compare these two orthologues. We revealed that their putative binding pockets differed by only three amino acids, and that RpalOR1 response spectrum mirrored that of RferOR1. Notably, RpalOR1 was strongly activated by the two components of the R. ferrugineus aggregation pheromone. Dose-response experiments further confirmed this functional conservation. Interestingly, we also found that both receptors were not specifically tuned to pheromones as they were also activated by high doses of host plant volatiles.
2. Material and methods
2.1. RpalOR1 and RferOR1 3D structure modelling and comparison of amino acids in the putative binding pockets
Five relaxed models were constructed with AlphaFold2 (Jumper et al., 2021) based on amino acid sequences of RpalOR1 (GenBank accession number QTG40723.1) and RferOR1 (QCS37752.1). For each receptor, the model with the highest pLDDT score was selected. The alignment between the sequences of RpalOR1 and RferOR1 was done using the MAFFT online service (Katoh et al., 2019). Detection of all RferOR1 and RpalOR1 cavities was performed using fpocket 3.0 (Le Guilloux et al., 2009) with default settings. The Cryo-EM structures of the complex MhraOR5-eugenol and MhraOR5-DEET (Protein Data Bank accession 7LID and 7LIG) were aligned with the RpalOR1 and RferOR1 3D models on PyMOL 2.5.4 (DeLano, 2002). After visually inspecting all the generated cavities, the main cavity situated at the same location as the binding site of MhraOR5 was identified as the putative binding site of each RpalOR1 and RferOR1. All residues within 5 Ångströms from the centre of these cavities were considered as constituents of the binding sites.
2.2. UAS-RpalOR1 construct and expression in Drosophila melanogaster
Generation of the UAS-RferOR1 fly line was described previously (Antony et al., 2021). For RpalOR1, the open reading frame (Genbank accession number MT887347.1) was synthesised in vitro by GenScript (Piscataway, NJ, USA) and subcloned into the pUAST.attB vector. The pUAST.attB-RpalOR1 plasmid was injected into Drosophila embryos with the genotype y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP}ZH-51C by BestGene Inc. (Chino Hills, CA, USA), allowing the use of the PhiC31 integrase system. This resulted in inserting the UAS-RpalOR1 construct into the genomic locus 51C of the second chromosome. UAS-RferOR1 and UAS-RpalOR1 lines were crossed with the Or67dGAL4 line to obtain homozygous flies expressing the transgene in at1 OSNs instead of OR67d. Flies were reared on a standard diet of cornmeal-yeast-agar medium and maintained in a climate- and light-controlled environment (25 °C, 12 h light: 12 h dark cycle).
2.3. Chemicals
Hexane and paraffin oil were purchased from Carlo Erba Reagents (Val de Reuil, France). The panel of beetle pheromones and structurally related compounds used in electrophysiology experiments was the same as previously used for the functional characterization of RferOR1 (Antony et al., 2021), except that 3-methyloctan-4-ol (phoenicol), the aggregation pheromone of R. phoenicis (Gries et al., 1993), was also included. The panel of host plant volatiles was composed of 26 compounds (Table 1) chosen because they are emitted by host plants of R. palmarum and R. ferrugineus and/or highly active on R. ferrugineus antennae (Abbas et al., 2006; Gries et al., 1994; Guarino et al., 2011; Jaffé et al., 1993; Rochat et al., 2000; Vacas et al., 2014). As preliminary experiments showed a strong activity of esters, we also enriched the panel with 12 supplementary ester molecules. Plant volatiles were purchased from Sigma-Aldrich (Saint-Louis, MO, USA), except anisole, ethyl caprylate, 2-methyl 1-butanol and methyl isobutyrate from Honeywell Fluka (Charlotte, NC, USA) and ethyl tiglate from Lancaster Synthesis (Morecambe, UK).
Table 1.
List of host plant volatiles and structurally related molecules used for stimulation of Drosophila OSNs expressing RpalOR1 and RferOR1.
| Chemical | Purity | CAS | InChiKey | Detection by R. ferrugineus antennae | Source of emission | References |
|---|---|---|---|---|---|---|
| acetophenone | 99% | 98–86–2 | KWOLFJPFCHCOCG-UHFFFAOYSA-N | yes | Phoenix dactylifera fruits | (1) (6) |
| 2-phenylethanol | 99% | 60–12–8 | WRMNZCZEMHIOCP-UHFFFAOYSA-N | yes | Elaeis guineensis fermented sap; Cocos nucifera crown; Phoenix canariensis affected stem; P. dactylifera fruits; Jacaratia digitata trunk; Saccharum officinarum stalk | (1) (2) (6) |
| methyl salicylate | >99% | 119–36–8 | OSWPMRLSEDHDFF-UHFFFAOYSA-N | yes | P. canariensis healthy tissue | (1) |
| methyl benzoate | 99% | 93–58–3 | QPJVMBTYPHYUOC-UHFFFAOYSA-N | yes | P. canariensis healthy tissue | (1) |
| anisole | >99% | 100–66–3 | RDOXTESZEPMUJZ-UHFFFAOYSA-N | yes | P. canariensis fermented leaves; J. digitata trunk | (1) (2) |
| nonanal | 95% | 124–19–6 | GYHFUZHODSMOHU-UHFFFAOYSA-N | yes | P. canariensis inflorescence; P. dactylifera fruits | (1) (6) |
| ethyl caprylate | >98% | 106–32–1 | YYZUSRORWSJGET-UHFFFAOYSA-N | yes | P. canariensis affected stem and fermented leaves; C. nucifera crown; E. guineensis fermented sap | (1) (2) |
| sulcatone | 99% | 110–93–0 | UHEPJGULSIKKTP-UHFFFAOYSA-N | yes | P. dactylifera fruits; P. canariensis affected stem and fermented leaves; S. officinarum stalk | (1) (2) (6) |
| hexanal | 95% | 66–25–1 | JARKCYVAAOWBJS-UHFFFAOYSA-N | yes | fermented C. nucifera apical part; P. canariensis inflorescence; P. dactylifera fruits; J. digitata trunk | (1) (2) (3) (6) |
| ethyl hexanoate | >99% | 123–66–0 | SHZIWNPUGXLXDT-UHFFFAOYSA-N | yes |
E. guineensis fermented sap; C. nucifera crown; P. canariensis affected stem and fermented leaves |
(1) (2) |
| 1-pentanol | 99% | 71–41–0 | AMQJEAYHLZJPGS-UHFFFAOYSA-N | yes | P. canariensis affected stem and fermented leaves | (1) |
| ethyl tiglate | 98% | 5837–78–5 | OAPHLAAOJMTMLY-GQCTYLIASA-N | C. nucifera crown | (2) | |
| ethyl valerate | 99% | 539–82–2 | ICMAFTSLXCXHRK-UHFFFAOYSA-N | yes | P. canariensis affected stem | (1) |
| ethyl isovalerate | 98% | 108–64–5 | PPXUHEORWJQRHJ-UHFFFAOYSA-N | C. nucifera crown; S. officinarum stalk | (2) | |
| methyl 2-methylbutyrate | >98% | 868–57–5 | OCWLYWIFNDCWRZ-UHFFFAOYSA-N | |||
| ethyl 2-methylbutyrate | >99% | 7452–79–1 | HCRBXQFHJMCTLF-UHFFFAOYSA-N | yes | P. canariensis affected stem | (1) |
| 2-methyl 1-butanol | >98% | 137–32–6 | QPRQEDXDYOZYLA-UHFFFAOYSA-N | yes | C. nucifera crown; E. guineensis fermented sap; S. officinarum stalk | (1) (2) |
| ethyl isobutyrate | >98% | 97–62–1 | WDAXFOBOLVPGLV-UHFFFAOYSA-N | yes | E. guineensis fermented sap, trunk; S. officinarum stalk | (1) (2) (4) (5) |
| methyl butyrate | 99% | 623–42–7 | UUIQMZJEGPQKFD-UHFFFAOYSA-N | yes | (1) | |
| methyl isobutyrate | >99% | 547–63–7 | BHIWKHZACMWKOJ-UHFFFAOYSA-N | |||
| ethyl butyrate | 99% | 105–54–4 | OBNCKNCVKJNDBV-UHFFFAOYSA-N | yes | C. nucifera crown, E. guineensis fermented sap, trunk; P. canariensis affected stem; S. officinarum stalk | (1) (2) (4) (5) |
| propyl butyrate | >95% | 105–66–8 | HUAZGNHGCJGYNP-UHFFFAOYSA-N | yes | P. canariensis affected stem | (1) |
| isopropyl butyrate | 99% | 638–11–9 | FFOPEPMHKILNIT-UHFFFAOYSA-N | |||
| butyl butyrate | 98% | 109–21–7 | XUPYJHCZDLZNFP-UHFFFAOYSA-N | yes | P. canariensis affected stem | (1) |
| isobutyl isobutyrate | 99% | 97–85–8 | RXGUIWHIADMCFC-UHFFFAOYSA-N | |||
| ethyl 3-hydroxybutyrate | 99% | 5405–41–4 | OMSUIQOIVADKIM-UHFFFAOYSA-N | |||
| butyric acid | 99% | 107–92–6 | FERIUCNNQQJTOY-UHFFFAOYSA-N | yes | P. canariensis affected and fermented stem | (1) |
| ethyl propionate | 99% | 105–37–3 | FKRCODPIKNYEAC-UHFFFAOYSA-N | yes | E. guineensis trunk; P. canariensis affected stem, fermented leaves; S. officinarum stalk | (1) (2) (4) (5) (7) |
| isobutyl propionate | >98% | 540–42–1 | FZXRXKLUIMKDEL-UHFFFAOYSA-N | E. guineensis trunk; S. officinarum stalk | (2) (4) | |
| propyl propionate | >99% | 106–36–5 | MCSINKKTEDDPNK-UHFFFAOYSA-N | yes | (1) | |
| isoamyl propionate | >98% | 105–68–0 | XAOGXQMKWQFZEM-UHFFFAOYSA-N | yes | P. canariensis affected stem | (1) |
| methyl acetate | >99.8% | 79–20–9 | KXKVLQRXCPHEJC-UHFFFAOYSA-N | |||
| ethyl acetate | 99.8% | 141–78–6 | XEKOWRVHYACXOJ-UHFFFAOYSA-N | yes | P. canariensis healthy and affected stem, fermented leaves; P. dactylifera fruits; S. officinarum stalk; | (1) (2) (4) (5) (6) (7) |
| sec-butyl acetate | 99% | 105–46–4 | DCKVNWZUADLDEH-UHFFFAOYSA-N | yes | P. canariensis affected stem | (1) |
| isobutyl acetate | 99% | 110–19–0 | GJRQTCIYDGXPES-UHFFFAOYSA-N | P. canariensis affected stem and fermented leaves; C. nucifera crown; S. officinarum stalk | (1) (2) | |
| propyl acetate | 99% | 109–60–4 | YKYONYBAUNKHLG-UHFFFAOYSA-N | E. guineensis fermented sap; S. officinarum stalk | (2) | |
| isopropyl acetate | 98% | 108–21–4 | JMMWKPVZQRWMSS-UHFFFAOYSA-N | |||
| acetic acid | 98% | 64–19–7 | QTBSBXVTEAMEQO-UHFFFAOYSA-N | yes | E. guineensis fermented sap; P. canariensis affected stem; P. dactylifera fruits; S. officinarum stalk | (1) (2) (6) |
2.4. Single-sensillum recordings
Single-sensillum recordings were carried out following the methods previously described (de Fouchier et al., 2015). at1 OSNs were subjected to 500 ms odorant stimulation using Pasteur pipette cartridges loaded with an odorant solution deposited onto a 1 cm2 filter paper. For screening experiments, cartridges were loaded with 1 µg of a pheromone compound diluted in 10 µL of hexane or 100 µg of a plant volatile diluted in 10 µL of paraffin oil. Cartridges containing hexane, paraffin oil and 10 µg of cis-vaccenyl acetate (to verify the absence of OR67d) were used as controls. For the comparison of dose–response curves between RpalOR1 and RferOR1, odorant doses ranged from 1 ng to 10 μg for pheromone compounds (diluted in 10 µL of hexane) and from 1 ng to 100 µg for plant volatiles (diluted in 10 µL of paraffin oil). For the comparison of dose-response curves of RpalOR1 to both pheromone compounds and plant volatiles, doses ranged from 1 ng to 100 µg and all chemicals were diluted in 10 µL of paraffin oil. Net responses of at1 OSNs expressing RpalOR1 or RferOR1 were computed by subtracting the spontaneous firing rate measured during 500 ms before stimulation from the firing rate measured during the 500 ms of odorant stimulation. Data were analysed with R (version 4.2.2). The normal distribution of the data was tested with a Shapiro-Wilk test. Since data did not follow a normal distribution, OSN responses to odorant stimulations were compared using a Kruskal-Wallis test, followed either by a Dunnett's multiple comparison test with a Benjamini & Hochberg correction in order to compare odorant responses to the control, or by a Dunn's multiple comparison test with a Benjamini & Hochberg correction in order to compare responses to different doses between RferOR1 and RpalOR1.
3. Results
3.1. 3D structure modelling of RpalOR1 and RferOR1: a strong structural similarity suggests a similar function
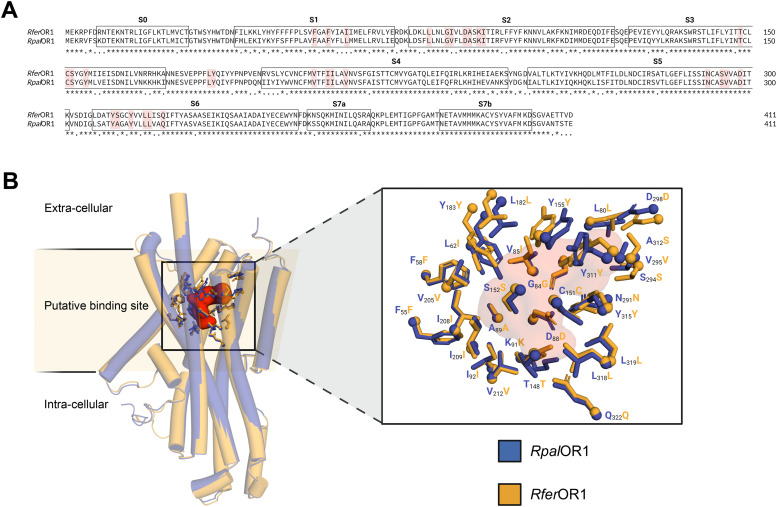
We first compared the 3D structure models of RpalOR1 and RferOR1 as determined by AlphaFold2. These two models exhibited a high similarity in their transmembrane segments (RMSD=1.28 Å), correlated with a global sequence identity of over 82 % (Fig. 1A,B). More than 30 cavities were identified using fpocket across all the models. Superimposing the RpalOR1 and RferOR1 models onto the experimental structures of MhraOR5 (del Mármol et al., 2021) allowed us to determine a cavity whose position was similar to the ligand binding site of MhraOR5, and which was delimited by 30 amino acids belonging to helices S2, 3, 4 and 6. Interestingly, these 30 amino acids were extremely conserved between RpalOR1 and RferOR1, with 90 % identity (Fig. 1A,B). Variations were exclusively detected at positions 62 (Leucine in RpalOR1 vs. Isoleucine in RferOR1), 85 (Valine vs. Isoleucine) and 312 (Alanine vs. Serine). The first two mutations probably have no significant impact on the structure and function of the binding site, as an amino acid with a hydrophobic side chain is replaced by another. Furthermore, the positioning of the residue 312 at the apex of the cavity could potentially mitigate its influence on the receptor's affinity for ligands, even in the context of substituting an Alanine (amino acid with a hydrophobic side chain) with a Serine (polar uncharged side chain). These results thus suggested a conservation of the response spectra of both receptors.
Fig. 1.
The predicted binding pockets of RpalOR1 and RferOR1 differ by only three amino acids (L62I, V85I and A312S). (A) Alignment and comparison of the two amino acid sequences (identity=82.24%). The helices (S) of both receptors are indicated by black boxes. Amino acids predicted to form the binding pocket are highlighted in red. (B) Visualisation of the binding pocket shared by both proteins and the amino acids that make it up. The assumed volume of the binding pocket is coloured red. RpalOR1 and RferOR1 are represented in blue and orange, respectively. The figure was created using the molecular visualisation software PyMOL.
3.2. RferOR1 and RpalOR1 are activated by the same pheromone compounds
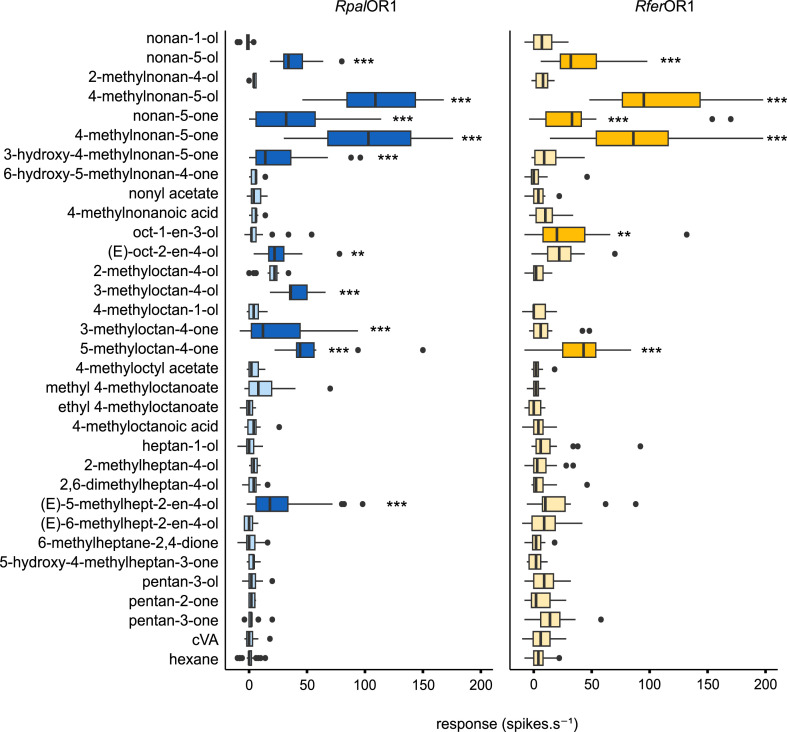
To verify whether the function of the orthologs RpalOR1 and RferOR1 was indeed conserved, we expressed RpalOR1 in Drosophila at1 neurons and stimulated these neurons with the panel of pheromone compounds and structural analogs previously used for the functional characterization of RferOR1 (Antony et al., 2021), plus 3-methyloctan-4-ol (phoenicol), the aggregation pheromone of the African palm weevil R. phoenicis (Gries et al., 1993). RpalOR1 was significantly activated by ten compounds, five of which are aggregation pheromones of palm weevils (Fig. 2). The best agonists were the two components of the R. ferrugineus aggregation pheromone. Ferrugineol (4-methylnonan-5-ol) and ferrugineone (4-methylnonan-5-one) elicited average responses of 112 and 103 action potentials per second, respectively. Additionally, RpalOR1 was activated by phoenicol as well as nonan-5-ol, 3‑hydroxy-4-methylnonan-5-one and 3-methyloctan-4-one, three of the five aggregation pheromone compounds of the West Indian sugarcane weevil Metamasius hemipterus (Ramirez-Lucas et al., 1996). We observed no response to the R. palmarum aggregation pheromone (E)−6-methylhept-2-en-4-ol (rhynchophorol), but its isomer (E)−5-methylhept-2-en-4-ol elicited an average response of 27 action potentials per second. We also found a response to 5-methyloctan-4-one, which is not known as an aggregation pheromone but is structurally similar to cruentol (5-methyloctan-4-ol), the aggregation pheromone of the palmetto weevil R. cruentatus (Weissling et al., 1994). The two last agonists were (E)‑oct-2-en-4-ol and nonan-5-one, which are also structurally related to aggregation pheromone compounds. Comparison of RpalOR1 response spectrum to that of RferOR1 (Antony et al., 2021) revealed a strong functional conservation between the two orthologues, even though a few moderate responses were found significant for one OR but not for the other (e.g. for oct‑1-en-3-ol and (E)‑oct-2-en-4-ol).
Fig. 2.
RpalOR1 and RferOR1 exhibit similar response spectra. Action potential frequency of Drosophila at1 OSNs expressing RpalOR1 or RferOR1 in response to beetle aggregation pheromones and structurally related molecules (1 µg loaded in the stimulus cartridge). Data for RferOR1 are from (Antony et al., 2021). Box plots show the median and the first and third quartiles of the distribution, whiskers show data distribution below the first quartile and above the third quartile, and dots show outliers. Dark blue and orange colours show significant differences from the solvent for RpalOR1 and RferOR1, respectively (Kruskal Wallis test followed by a Dunnett's multiple comparison test with a Benjamini & Hochberg correction, n = 11–26 for RpalOR1, n = 14–38 for RferOR1, *p < 0.05, ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001).
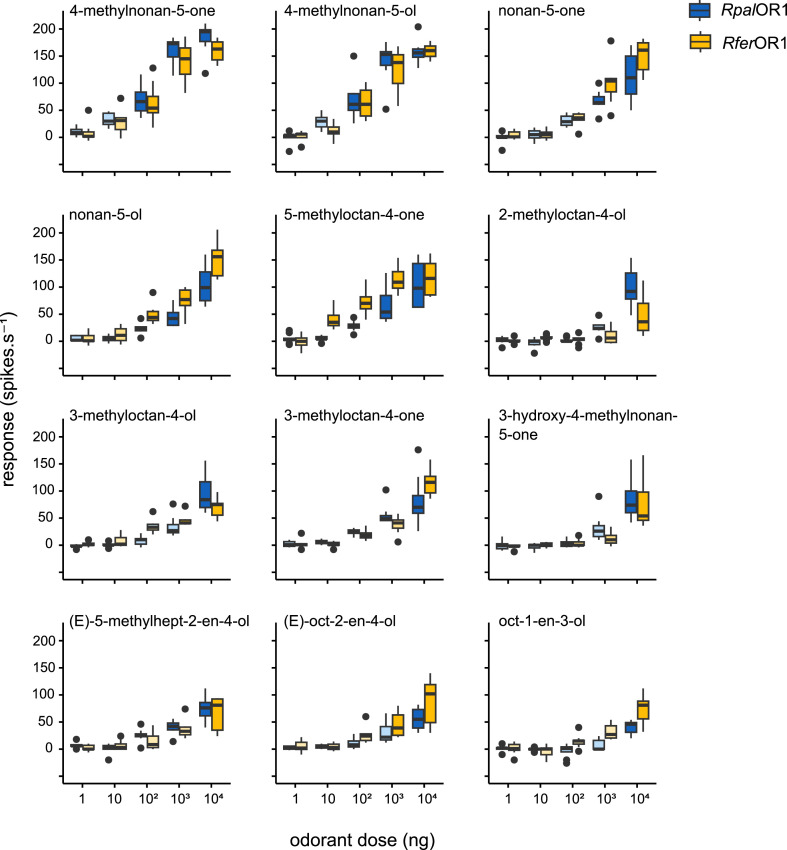
To further verify the functional conservation between the two receptors, we performed dose-response experiments on Drosophila neurons expressing RpalOR1 or RferOR1 with the 11 compounds that elicited significant responses at high dose, plus 2-methyloctan-4-ol for which small responses could be measured for RpalOR1, although not significant. For both receptors, the best two ligands ferrugineol and ferrugineone elicited responses significantly different from the solvent starting from a dose of 100 ng (Fig. 3, Supplementary Table S1). The response threshold was raised at 1 µg for nonan-5-one, nonan-5-ol and 5-methyloctan-4-one, and at 10 µg for the seven other odorants. For some compounds, the statistical analysis revealed a different response threshold for the two ORs. This was notably the case for 5-methyloctan-4-one, which elicited significant responses starting from 10 ng for RferOR1 and 1 µg for RpalOR1. However, we found no statistically significant difference when comparing responses of the two orthologs to the same dose of an odorant. Overall, dose-response experiments thus confirmed that RpalOR1 and RferOR1 have highly similar response spectra, although not fully identical.
Fig. 3.
RpalOR1 and RferOR1 have similar sensitivities to their ligands. Dose-response experiments performed on at1 OSNs expressing RpalOR1 or RferOR1 with the 12 best agonists. Box plots show the median and the first and third quartiles of the distribution, whiskers show data distribution below the first quartile and above the third quartile, and dots show outliers. Dark blue and orange colours show significant differences from the solvent for RpalOR1 and RferOR1, respectively (Kruskal Wallis test followed by a Dunnett's multiple comparison test with a Benjamini & Hochberg correction, p-values available in Supplementary Table S1). No significant difference was observed between RpalOR1 and RferOR1 responses to a given odorant dose (Kruskal-Wallis test followed by a Dunn's multiple comparison test with a Benjamini & Hochberg correction, n = 7–12, p > 0.05).
3.3. RpalOR1 and RferOR1 are not specifically tuned to pheromones as they also respond to host volatiles
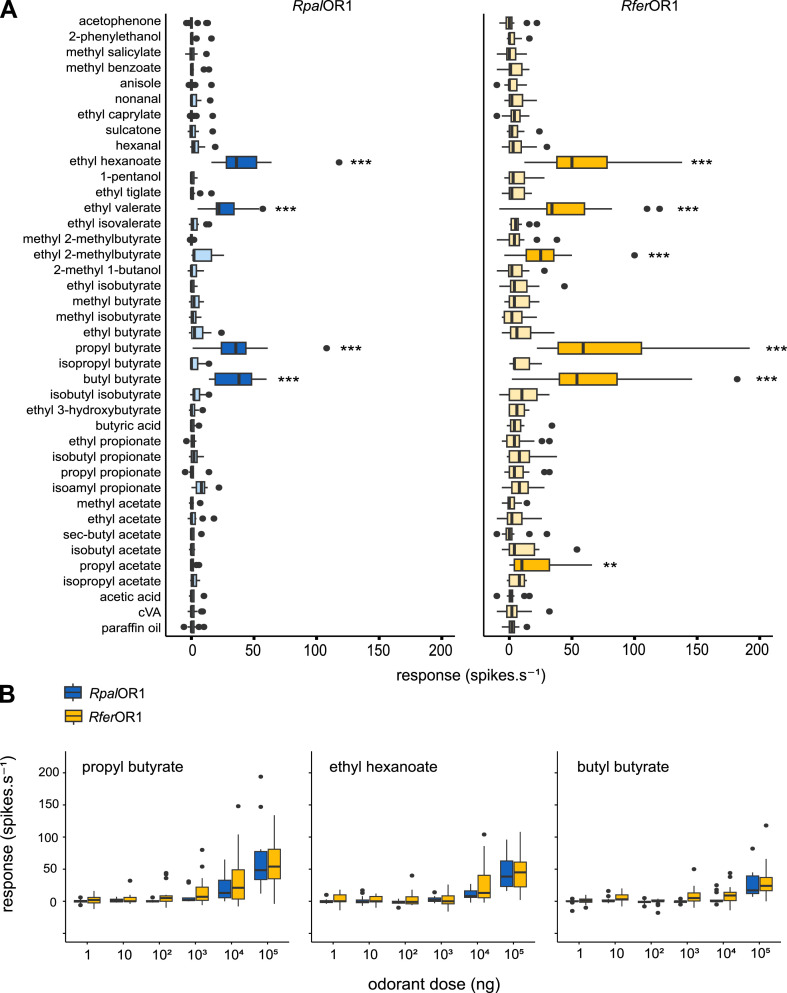
Since both RpalOR1 and RferOR1 exhibited a relatively broad odorant tuning, unusual for pheromone receptors, we next wondered whether they could also detect host plant volatiles. We selected volatile compounds previously shown to be emitted by R. palmarum and R. ferrugineus hosts and to be detected by R. ferrugineus antennae (Rochat et al., 2000; Vacas et al., 2014), and tested them at high dose on at1 OSNs expressing the pheromone receptors. This revealed that both RpalOR1 and RferOR1 were significantly activated by the same palm tree esters: propyl butyrate, butyl butyrate, ethyl valerate and ethyl hexanoate (Fig. 4A). Neurons expressing RferOR1 also significantly responded to ethyl-2-methylbutyrate and propyl acetate. Dose-response experiments for the three most potent ligands further showed identical response thresholds for the two receptors (Fig. 4B).
Fig. 4.
RpalOR1 and RferOR1 are activated by host plant volatiles. (A) Action potential frequency of Drosophila at1 OSNs expressing RpalOR1 or RferOR1 in response to palm tree volatiles and structurally related molecules (100 µg in the stimulus cartridge). Dark blue and orange colours show significant differences from the solvent (Kruskal-Wallis test followed by a Dunnett's multiple comparison test with a Benjamini & Hochberg correction, n = 12–38, *p < 0.05, ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001). (B) Dose-response experiments performed on at1 OSNs expressing RpalOR1 or RferOR1 with the three best agonists. Box plots show the median and the first and third quartiles of the distribution, whiskers show data distribution below the first quartile and above the third quartile, and dots show outliers. No significant difference was observed between RpalOR1 and RferOR1 responses to a given odorant dose (Kruskal-Wallis test followed by a Dunn's multiple comparison test with a Benjamini & Hochberg correction, n = 13–16, p > 0.05).
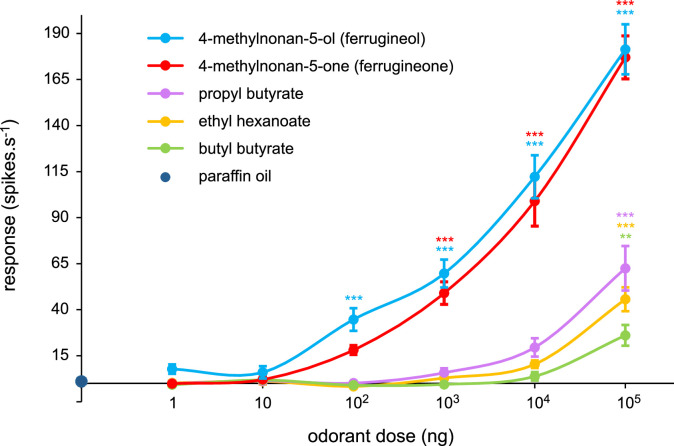
We next performed dose-response experiments in order to compare the sensitivity of RpalOR1 towards the two most active pheromone compounds (ferrugineol and ferrugineone) and the three most active host plant volatiles (propyl butyrate, ethyl hexanoate and butyl butyrate). To do so, all molecules were dissolved in the same solvent, i.e. paraffin oil. RpalOR1 was much more sensitive to ferrugineol and ferrugineone than to plant volatiles (Fig. 5). Significant responses were recorded starting from a dose of 100 ng of ferrugineol and 1 µg of ferrugineone vs. 100 µg of the palm esters propyl butyrate, ethyl hexanoate and butyl butyrate. At the highest dose, average responses to pheromones reached 181 and 177 action potentials per second for ferrugineol and ferrugineone, respectively, vs. 63 for propyl butyrate, the most potent host plant volatile.
Fig. 5.
RpalOR1 is more sensitive to R. ferrugineus pheromone compounds than to palm esters. Dose-response curves of Drosophila at1 OSNs expressing RpalOR1 to the two most active pheromone compounds and the three most active plant volatiles diluted in paraffin oil. These curves show mean action potential frequencies at different odorant doses +/- SEM (Kruskal-Wallis test followed by a Dunnett's multiple comparison test with a Benjamini & Hochberg correction, n = 10–27, ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001).
4. Discussion
The palm weevils R. ferrugineus and R. palmarum are currently ranked as the most devastating insect pests to palm trees, being responsible for millions of economic losses (Hoddle et al., 2024). Despite their different distribution areas, these two species are genetically closely related and present behavioural and ecological similarities. Males from both species produce an aggregation pheromone to attract males and females, although the chemical structures of their components differ (Hallett et al., 1993; Perez et al., 1996; Oehlschlager et al., 1992; Rochat et al., 1991). The recent sequencing of the R. palmarum antennal transcriptome revealed that both weevils harbour a similar repertoire of ORs (Gonzalez et al., 2021). Out of the 63 RpalORs identified in the transcriptome, at least 57 had a corresponding RferOR ortholog. Among these ORs, the amino acid sequence of RpalOR1 shared 82.24 % identity with its ortholog RferOR1. Since RferOR1 has been characterised as a receptor to the aggregation pheromone of R. ferrugineus, a first hypothesis would be that RpalOR1 is the receptor to the aggregation pheromone of R. palmarum, as initially proposed (Gonzalez et al., 2021). It is indeed possible that the 20 % amino acid differences alter the pheromone-binding site, making RpalOR1 responsible for detecting rhynchophorol instead of ferrugineol/ferrugineone in RferOR1. Alternatively, another hypothesis would be that RferOR1 and RpalOR1 detect similar compounds, especially if the amino acid differences are not in the binding site.
To test these hypotheses, we first reported here the 3D models of the two ORs using AlphaFold2, revealing that their potential pheromone-binding pockets were much conserved, arguing in favour of the second hypothesis. AlphaFold2 has recently shown remarkable success in predicting the 3D structures of a wide range of proteins, including membrane-bound receptors such as ORs. However, accuracy and performance of such a computational method can still be questioned for insect ORs, as only two experimental structures are yet available (Butterwick et al., 2018; del Mármol et al., 2021). Thus, we next conducted functional studies to assess RpalOR1 ability to bind pheromone compounds. The results confirmed the hypothesis of a conservation of response spectra, since RpalOR1 response profile to the different agonists perfectly matched that of RferOR1. Such an evolutionary conservation of OR tuning has already been reported in Curculionidae, in two lineages of orthologous ORs from the conifer-feeding bark beetles Ips typographus and Dendroctonus ponderosae, and the pine weevil Hylobius abietis (Guo et al., 2021; Roberts et al., 2022). In this latter study, the orthologous ORs responded to volatiles that are ecologically relevant to the three species, namely 2-phenylethanol and angiosperm green leaf volatiles. Interestingly, response spectra were highly conserved despite amino-acid sequence identities as low as 50 %, showing that overall sequence identity is a poor predictor of functional conservation. Here, we found no response of RpalOR1 to rhynchophorol - the aggregation pheromone of R. palmarum - but strong responses to ferrugineol and ferrugineone, which are the two components of the R. ferrugineus aggregation pheromone. These results show that RpalOR1, contrary to RferOR1, is not involved in intraspecific pheromone communication. Consequently, these two receptors have different biological functions although they share a similar tuning. RpalOR1 is one of the most highly expressed ORs in the antennae of R. palmarum, but there are two other receptors identified in the antennal transcriptome with even higher expression levels (Gonzalez et al., 2021). As high expression is often used as a criterion to identify pheromone receptors in insects (Fleischer and Krieger, 2018; Antony et al., 2021), these two ORs appear as good pheromone receptor candidates.
R. palmarum OSNs slightly responding to ferrugineol were identified previously (Saïd et al., 2003), confirming that this weevil can detect heterospecific pheromone compounds. However, these OSNs were also activated by rhynchophorol, suggesting that RpalOR1 may not be the OR expressed in these OSNs. To our knowledge, the detection of ferrugineone and the other RpalOR1-active weevil pheromones by the antennae of R. palmarum has not been tested. Finding an R. palmarum OR tuned to the aggregation pheromone of R. ferrugineus raises the question of the ecological relevance of this detection, as the two species do not have overlapping distribution areas. However, ferrugineol has also been reported as the major aggregation pheromone for the neotropical weevils M. hemipterus and Dynamis borassi (Giblin-Davis et al., 1997; Ramirez-Lucas et al., 1996). As these species are found in sympatry with R. palmarum, this compound may be used by R. palmarum to detect the presence of heterospecifics. Whether ferrugineol may act as a repellent or attraction inhibitor to R. palmarum remains to be experimentally tested. Alternatively, it has been proposed that aggregation pheromones, including ferrugineol, may be used as synomones by palm weevils (Bandeira et al., 2021; Giblin-Davis et al., 1996; Oehlschlager, 2016). Indeed, cross-attraction/cross-detection of aggregation pheromones has been frequently observed in weevils, particularly in neotropical palm weevils. For instance, aggregation pheromones of R. cruentatus, R. palmarum and R. phoenicis are detected by antennae of M. hemipterus (Ramirez-Lucas et al., 1996), and D. borassi, M. hemipterus and Placoclytus distortus have been reported to be attracted to rhynchophorol (Giblin-Davis et al., 1996). It has been proposed that such a cross-attraction of sympatric weevils may have evolved to optimise the search of food resources or to overcome host plant defence.
Interestingly, we found that RpalOR1 and RferOR1 were not specifically tuned to pheromones as they were also activated by four of the 38 plant volatiles tested. It is uncommon for a receptor to have the capacity to bind both pheromones and plant volatiles. However, the latter are typically not tested on pheromone receptors, so this co-detection may have been overlooked. Our results are anyhow in line with some studies conducted in other insects orders, which documented the activation of pheromone receptors by plant volatiles in moths (Yuvaraj et al., 2017; Zhang et al., 2024) and in the pea aphid (Zhang et al., 2017). The four odorants detected by RpalOR1 and RferOR1 (ethyl hexanoate, ethyl valerate, propyl butyrate and butyl butyrate) are esters known to be emitted by Canary Island date palm stems already infested by weevil larvae and they are detected by R. ferrugineus antennae (Vacas et al., 2014). It is challenging to ascertain the ecological relevance of the doses we identified as active (10 to 100 µg on the cartridge filter paper, dependent on the compound) to the insect. Firstly, the actual dose that reaches the Drosophila neurons in single-sensillum recordings is unknown, but it is likely to be considerably lower than the dose deposited on the filter paper. Secondly, data on the doses of volatiles emitted by palm trees are scarce, and are reported as quantity by volume and time. In any case, 0.5 to 10 mg.m−2.h−1 of compounds are typically emitted by palm trees, as measured by Proton-Transfer-Reaction Mass Spectrometry in a plantation in South East Asia (Misztal et al., 2011). Consequently, we can assume that the quantities used in the present study are of a comparable magnitude to those encountered by the insect in the field, and that the OR responses observed are unlikely to be the result of excessive dosage. However, the precise role of these esters in the ecology of palm weevils has not been investigated. In the context of improving pest management methods based on semiochemicals, it has been found on multiple occasions that attraction of palm weevils towards aggregation pheromone-baited traps is increased by the addition of fermenting host plant material (Bandeira et al., 2021; Giblin-Davis et al., 1996; Oehlschlager, 2016). This indicates that palm volatiles can act in synergy with pheromones to promote attraction but until now, only a few synthetic volatiles with such a synergistic effect have been identified, with contrasting results (Gries et al., 1994; Guarino et al., 2011; Rochat et al., 2000; Vacas et al., 2017, 2014, 2013). It would be of interest to verify the effect of RferOR1 ligands identified in the frame of this study on trap catches in the field, although synergistic effects are more likely to be mediated by other ORs specifically tuned to palm volatiles, such as the recently identified RferOR2 (Antony et al., 2024). It is also possible that the ability of OR1 orthologs to bind some plant volatiles has no adaptive value but is rather a trace of its descent from an ancestral OR tuned to host volatiles. This calls for a broader comparative study that would help to unravel the evolutionary history of these original pheromone receptors.
CRediT authorship contribution statement
Ludvine Brajon: Investigation, Formal analysis, Writing – original draft. Arthur Comte: Investigation, Formal analysis, Writing – original draft. Rémi Capoduro: Investigation, Writing – review & editing. Camille Meslin: Investigation, Writing – review & editing. Binu Antony: Conceptualization, Funding acquisition, Project administration, Resources, Writing – review & editing. Mohammed Ali Al-Saleh: Funding acquisition, Project administration. Arnab Pain: Funding acquisition, Project administration. Emmanuelle Jacquin-Joly: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft. Nicolas Montagné: Conceptualization, Project administration, Supervision, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by Sorbonne Université, INRAE, KSU, KAUST (grants KAUST-OSR-2018-RPW-3816-1 and OSR-2018-RPW-3816–4) and by the French program “Cultiver et Protéger Autrement” (grant ANR-20-PCPA-0007). The authors extend their appreciation to the Deanship of Scientific Research, King Saud University, for funding through the Vice Deanship of Scientific Research Chairs, Chair of Date Palm Research.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cris.2024.100090.
Contributor Information
Emmanuelle Jacquin-Joly, Email: emmanuelle.joly@inrae.fr.
Nicolas Montagné, Email: nicolas.montagne@sorbonne-universite.fr.
Appendix. Supplementary materials
Data availability
Raw data are available in Supplementary Table S2.
References
- Abbas M.S.T., Hanounik S.B., Shahdad A.S., AI-Bagham S.A. Aggregation pheromone traps, a major component of IPM strategy for the red palm weevil, Rhynchophorus ferrugineus in date palms (Coleoptera: Curculionidae) J. Pest Sci. 2006;79:69–73. doi: 10.1007/s10340-005-0113-6. [DOI] [Google Scholar]
- Antony B., Johny J., Montagné N., Jacquin-Joly E., Capoduro R., Cali K., Persaud K., Al-Saleh M.A., Pain A. Pheromone receptor of the globally invasive quarantine pest of the palm tree, the red palm weevil (Rhynchophorus ferrugineus) Mol. Ecol. 2021;30:2025–2039. doi: 10.1111/mec.15874. [DOI] [PubMed] [Google Scholar]
- Antony B., Montagné N., Comte A., Mfarrej S., Jakše J., Capoduro R., Shelke R., Cali K., Al-Saleh M.A., Persaud K., Pain A., Jacquin-Joly E. Deorphanizing an odorant receptor tuned to palm tree volatile esters in the Asian palm weevil sheds light on the mechanisms of palm tree selection. Insect Biochem. Mol. Biol. 2024;169:104129. doi: 10.1016/j.ibmb.2024.104129. [DOI] [PubMed] [Google Scholar]
- Baldwin M.W., Ko M.-C. Functional evolution of vertebrate sensory receptors. Horm. Behav. 2020;124 doi: 10.1016/j.yhbeh.2020.104771. [DOI] [PubMed] [Google Scholar]
- Bandeira P.T., Fávaro C.F., Francke W., Bergmann J., Zarbin P.H.G. Aggregation Pheromones of Weevils (Coleoptera: Curculionidae): Advances in the Identification and Potential Uses in Semiochemical-Based Pest Management Strategies. J. Chem. Ecol. 2021;47:968–986. doi: 10.1007/s10886-021-01319-1. [DOI] [PubMed] [Google Scholar]
- Benton R., Sachse S., Michnick S.W., Vosshall L.B. Atypical Membrane Topology and Heteromeric Function of Drosophila Odorant Receptors In Vivo. PLOS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterwick J.A., del Mármol J., Kim K.H., Kahlson M.A., Rogow J.A., Walz T., Ruta V. Cryo-EM structure of the insect olfactory receptor Orco. Nature. 2018;560:447–452. doi: 10.1038/s41586-018-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Liu Y., Wang B., Wang G. A single point mutation causes one-way alteration of pheromone receptor function in two Heliothis species. iScience. 2021;24 doi: 10.1016/j.isci.2021.102981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Shi C., Wang B., Xiu P., Wang Y., Liu Y., Wang G. Evolutionary shifts in pheromone receptors contribute to speciation in four Helicoverpa species. Cell. Mol. Life Sci. 2023;80:199. doi: 10.1007/s00018-023-04837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fouchier A., Sun X., Monsempes C., Mirabeau O., Jacquin-Joly E., Montagné N. Evolution of two receptors detecting the same pheromone compound in crop pest moths of the genus Spodoptera. Front. Ecol. Evol. 2015;3 doi: 10.3389/fevo.2015.00095. [DOI] [Google Scholar]
- del Mármol J., Yedlin M.A., Ruta V. The structural basis of odorant recognition in insect olfactory receptors. Nature. 2021;597:126–131. doi: 10.1038/s41586-021-03794-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. PyMOL : An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr. 2002;40:82–92. [Google Scholar]
- Fleischer J., Krieger J. Insect Pheromone Receptors – Key Elements in Sensing Intraspecific Chemical Signals. Front. Cell. Neurosci. 2018;12:425. doi: 10.3389/fncel.2018.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers J.M., Hazzouri K.M., Lemansour A., Capote T., Gros-Balthazard M., Ferrand S., Lebrun M., Amiri K.M.A., Purugganan M.D. Patterns of Volatile Diversity Yield Insights Into the Genetics and Biochemistry of the Date Palm Fruit Volatilome. Front. Plant Sci. 2022;13:853651. doi: 10.3389/fpls.2022.853651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber K., Giblin-Davis R.M. Association of the Red Ring Nematode and Other Nematode Species with the Palm Weevil, Rhynchophorus palmarum. J. Nematol. 1990;22:143–149. [PMC free article] [PubMed] [Google Scholar]
- Giblin-Davis R.M., Gries R., Gries G., Peña-Rojas E., Pinzón I., Peña J.E., Perez A.L., Pierce H.D., Oehlschlager A.C. Aggregation Pheromone of Palm Weevil, Dynamis borassi. J. Chem. Ecol. 1997;23:2287–2297. doi: 10.1023/B:JOEC.0000006674.64858.f2. [DOI] [Google Scholar]
- Giblin-Davis R.M., Oehlschlager A.C., Perez A., Gries G., Gries R., Weissling T.J., Chinchilla C.M., Peña J.E., Hallett R.H., Pierce H.D., Gonzalez L.M. Chemical and Behavioral Ecology of Palm Weevils (Curculionidae: Rhynchophorinae) Fla. Entomol. 1996;79:153–167. doi: 10.2307/3495812. [DOI] [Google Scholar]
- Gonzalez F., Johny J., Walker W.B., Guan Q., Mfarrej S., Jakše J., Montagné N., Jacquin-Joly E., Alqarni A.S., Al-Saleh M.A., Pain A., Antony B. Antennal transcriptome sequencing and identification of candidate chemoreceptor proteins from an invasive pest, the American palm weevil Rhynchophorus palmarum. Sci. Rep. 2021;11:8334. doi: 10.1038/s41598-021-87348-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gries G., Gries R., Perez A.L., Gonzales L.M., Pierce H.D., Oehlschlager A.C., Rhainds M., Zebeyou M., Kouame B. Ethyl propionate: Synergistic kairomone for african palm weevil, Rhynchophorus phoenicis L. (Coleoptera: Curculionidae) J. Chem. Ecol. 1994;20:889–897. doi: 10.1007/BF02059585. [DOI] [PubMed] [Google Scholar]
- Gries G., Gries R., Perez A.L., Oehlschlager A.C., Gonzales L.M., Pierce H.D., Kouda-Bonafos M., Zebeyou M., Nanou N. Aggregation pheromone of the African palm weevil. Rhynchophorus phoenicis F. Naturwissenschaften. 1993:90–91. doi: 10.1007/BF01140426. [DOI] [Google Scholar]
- Guarino S., Bue P.L., Peri E., Colazza S. Responses of Rhynchophorus ferrugineus adults to selected synthetic palm esters: electroantennographic studies and trap catches in an urban environment. Pest Manag. Sci. 2011;67:77–81. doi: 10.1002/ps.2035. [DOI] [PubMed] [Google Scholar]
- Guo M., Du L., Chen Q., Feng Y., Zhang J., Zhang X., Tian K., Cao S., Huang T., Jacquin-Joly E., Wang G., Liu Y. Odorant Receptors for Detecting Flowering Plant Cues Are Functionally Conserved across Moths and Butterflies. Mol. Biol. Evol. 2021;38:1413–1427. doi: 10.1093/molbev/msaa300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett R., Gries G., Gries R., Borden J.H., Czyzewska E., Oehlschlager A.C., Pierce H.D., Angerilli N., Rauf A. Aggregation pheromones of two asian palm Weevils, Rhynchophorus ferrugineus and R. vulneratus. Naturwissenschaften. 1993;80:328–331. doi: 10.1007/BF01141908. [DOI] [Google Scholar]
- Hoddle M., Antony B., El-Shafie H., Chamorro L., Milosavljević I., Löhr B., Faleiro R. Taxonomy, Biology, Symbionts, Omics, and Management of Rhynchophorus Palm Weevils (Coleoptera: Curculionidae: Dryophthorinae) Annual Review of Entomology. 2024;69:455–479. doi: 10.1146/annurev-ento-013023-121139. [DOI] [PubMed] [Google Scholar]
- Hou X.-Q., Yuvaraj J.K., Roberts R.E., Zhang D.-D., Unelius C.R., Löfstedt C., Andersson M.N. Functional Evolution of a Bark Beetle Odorant Receptor Clade Detecting Monoterpenoids of Different Ecological Origins. Mol. Biol. Evol. 2021;38:4934–4947. doi: 10.1093/molbev/msab218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffé K., Sánchez P., Cerda H., Hernández J.V., Jaffé R., Urdaneta N., Guerra G., Martínez R., Miras B. Chemical ecology of the palm weevil Rhynchophorus palmarum (L.) (Coleoptera: Curculionidae): Attraction to host plants and to a male-produced aggregation pheromone. J. Chem. Ecol. 1993;19:1703–1720. doi: 10.1007/BF00982302. [DOI] [PubMed] [Google Scholar]
- Ji T., Xu Z., Jia Q., Wang G., Hou Y. Non-palm Plant Volatile α-Pinene Is Detected by Antenna-Biased Expressed Odorant Receptor 6 in the Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae) Front. Physiol. 2021;12:701545. doi: 10.3389/fphys.2021.701545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinformatics. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary G.P., Allen J.E., Bunger P.L., Luginbill J.B., Linn C.E., Macallister I.E., Kavanaugh M.P., Wanner K.W. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. 2012;109:14081–14086. doi: 10.1073/pnas.1204661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Capoduro R., Bastin–Héline L., Zhang S., Sun D., Lucas P., Dabir-Moghaddam D., François M.-C., Liu Y., Wang G., Jacquin-Joly E., Montagné N., Meslin C. A tale of two copies: Evolutionary trajectories of moth pheromone receptors. Proc. Natl. Acad. Sci. 2023;120 doi: 10.1073/pnas.2221166120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misztal P.K., Nemitz E., Langford B., Di Marco C.F., Phillips G.J., Hewitt C.N., MacKenzie A.R., Owen S.M., Fowler D., Heal M.R., Cape J.N. Direct ecosystem fluxes of volatile organic compounds from oil palms in South-East Asia. Atmos. Chem. Phys. 2011;11:8995–9017. doi: 10.5194/acp-11-8995-2011. [DOI] [Google Scholar]
- Oehlschlager A.C. Palm Weevil Pheromones – Discovery and Use. J. Chem. Ecol. 2016;42:617–630. doi: 10.1007/s10886-016-0720-0. [DOI] [PubMed] [Google Scholar]
- Oehlschlager A.C., Chinchilla C.M., Gonzalez L.M., Jiron L.F., Mexzon R., Morgan B. Development of a Pheromone-Based Trapping System for Rhynchophorus palmarum (Coleoptera: Curculionidae) J. Econ. Entomol. 1993;86:1381–1392. doi: 10.1093/jee/86.5.1381. [DOI] [Google Scholar]
- Oehlschlager A.C., Pierce H.D., Morgan B., Wimalaratne P.D.C., Slessor K.N., King G.G.S., Gries G., Gries R., Borden J.H., Jiron L.F., Chinchilla C.M., Mexzan R.G. Chirality and field activity of rhynchophorol, the aggregation pheromone of the American palm weevil. Naturwissenschaften. 1992;79:134–135. doi: 10.1007/BF01131543. [DOI] [Google Scholar]
- Perez A.L., Hallett R.H., Gries R., Gries G., Oehlschlager A.C., Borden J.H. Pheromone chirality of asian palm weevils, Rhynchophorus ferrugineus (Oliv.) and R. vulneratus (Panz.) (Coleoptera: Curculionidae) J. Chem. Ecol. 1996;22:357–368. doi: 10.1007/BF02055104. [DOI] [PubMed] [Google Scholar]
- Ramirez-Lucas P., Malosse C., Ducrot P.-H., Lettere M., Zagatti P. Chemical identification, electrophysiological and behavioral activities of the pheromone of Metamasius hemipterus (Coleoptera: Curculionidae) Bioorg. Med. Chem. 1996;4:323–330. doi: 10.1016/0968-0896(96)00009-0. [DOI] [PubMed] [Google Scholar]
- Roberts R.E., Biswas T., Yuvaraj J.K., Grosse-Wilde E., Powell D., Hansson B.S., Löfstedt C., Andersson M.N. Odorant receptor orthologues in conifer-feeding beetles display conserved responses to ecologically relevant odours. Mol. Ecol. 2022;31:3693–3707. doi: 10.1111/mec.16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H.M. Molecular Evolution of the Major Arthropod Chemoreceptor Gene Families. Annu. Rev. Entomol. 2019;64:227–242. doi: 10.1146/annurev-ento-020117-043322. [DOI] [PubMed] [Google Scholar]
- Rochat D., Malosse C., Lettere M., Ducrot P.-H., Zagatti P., Renou M., Descoins C. Male-produced aggregation pheromone of the american palm weevil, Rhynchophorus palmarum (L.) (Coleoptera, Curculionidae): Collection, identification, electrophysiogical activity, and laboratory bioassay. J. Chem. Ecol. 1991;17:2127–2141. doi: 10.1007/BF00987996. [DOI] [PubMed] [Google Scholar]
- Rochat D., Nagnan-Le Meillour P., Esteban-Duran J., Malosse C., Perthuis B., Morin jean paul, Descoins C. Identification of Pheromone Synergists in American Palm Weevil, Rhynchophorus Palmarum, and Attraction of Related Dynamis Borassi. J. Chem. Ecol. 2000;26:155–187. doi: 10.1023/A:1005497613214. [DOI] [Google Scholar]
- Saïd I., Tauban D., Renou M., Mori K., Rochat D. Structure and function of the antennal sensilla of the palm weevil Rhynchophorus palmarum (Coleoptera, Curculionidae) J. Insect Physiol. 2003;49:857–872. doi: 10.1016/S0022-1910(03)00137-9. [DOI] [PubMed] [Google Scholar]
- Suh E., Bohbot J.D., Zwiebel L.J. Peripheral olfactory signaling in insects. Curr. Opin. Insect Sci. 2014;6:86–92. doi: 10.1016/j.cois.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacas S., Abad-Payá M., Primo J., Navarro-Llopis V. Identification of Pheromone Synergists for Rhynchophorus ferrugineus Trapping Systems from Phoenix canariensis Palm Volatiles. J. Agric. Food Chem. 2014;62:6053–6064. doi: 10.1021/jf502663y. [DOI] [PubMed] [Google Scholar]
- Vacas S., Melita O., Michaelakis A., Milonas P., Minuz R., Riolo P., Abbass M.K., Lo Bue P., Colazza S., Peri E., Soroker V., Livne Y., Primo J., Navarro-Llopis V. Lures for red palm weevil trapping systems: aggregation pheromone and synthetic kairomone. Pest Manag. Sci. 2017;73:223–231. doi: 10.1002/ps.4289. [DOI] [PubMed] [Google Scholar]
- Vacas S., Primo J., Navarro-Llopis V. Advances in the Use of Trapping Systems for Rhynchophorus ferrugineus (Coleoptera: Curculionidae): Traps and Attractants. J. Econ. Entomol. 2013;106:1739–1746. doi: 10.1603/EC13105. [DOI] [PubMed] [Google Scholar]
- Vizueta J., Escuer P., Frías-López C., Guirao-Rico S., Hering L., Mayer G., Rozas J., Sánchez-Gracia A. Evolutionary History of Major Chemosensory Gene Families across Panarthropoda. Mol. Biol. Evol. 2020;37:3601–3615. doi: 10.1093/molbev/msaa197. [DOI] [PubMed] [Google Scholar]
- Weissling T.J., Giblin-Davis R.M., Gries G., Gries R., Perez A.L., Pierce H.D., Oehlschlager A.C. Aggregation pheromone of palmetto weevil, Rhynchophorus cruentatus (F.) (Coleoptera: Curculionidae) J. Chem. Ecol. 1994;20:505–515. doi: 10.1007/BF02059593. [DOI] [PubMed] [Google Scholar]
- Yang K., Huang L.-Q., Ning C., Wang C.-Z. Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. eLife. 2017;6:e29100. doi: 10.7554/eLife.29100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaraj J.K., Roberts R.E., Sonntag Y., Hou X.-Q., Grosse-Wilde E., Machara A., Zhang D.-D., Hansson B.S., Johanson U., Löfstedt C., Andersson M.N. Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. BMC Biol. 2021;19:16. doi: 10.1186/s12915-020-00946-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaraj J.K., Corcoran J.A., Andersson M.N., Newcomb R.D., Anderbrant O., Löfstedt C. Characterization of Odorant Receptors from a Non-Ditrysian Moth, Eriocrania Semipurpurella Sheds Light on the Origin of Sex Pheromone Receptors in Lepidoptera. Mol Biol Evol. 2017;34:2733–2746. doi: 10.1093/molbev/msx215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang B., Grossi G., Falabella P., Liu Y., Yan S., Lu J., Xi J., Wang G. Molecular Basis of Alarm Pheromone Detection in Aphids. Curr Biol. 2017;27:55–61. doi: 10.1016/j.cub.2016.10.013. [DOI] [PubMed] [Google Scholar]
- Zhang S., Jacquin-Joly E., Montagné N., Liu F., Liu Y., Wang G. Identification of an odorant receptor responding to sex pheromones in Spodoptera frugiperda extends the novel type-I PR lineage in moths. Insect Sci. 2024;31:489–502. doi: 10.1111/1744-7917.13248. [DOI] [PubMed] [Google Scholar]
- Zhao Z., McBride C.S. Evolution of olfactory circuits in insects. J. Comp. Physiol. A. 2020;206:353–367. doi: 10.1007/s00359-020-01399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data are available in Supplementary Table S2.