Figure 5. Structural characterization of GP38-specific antibodies.
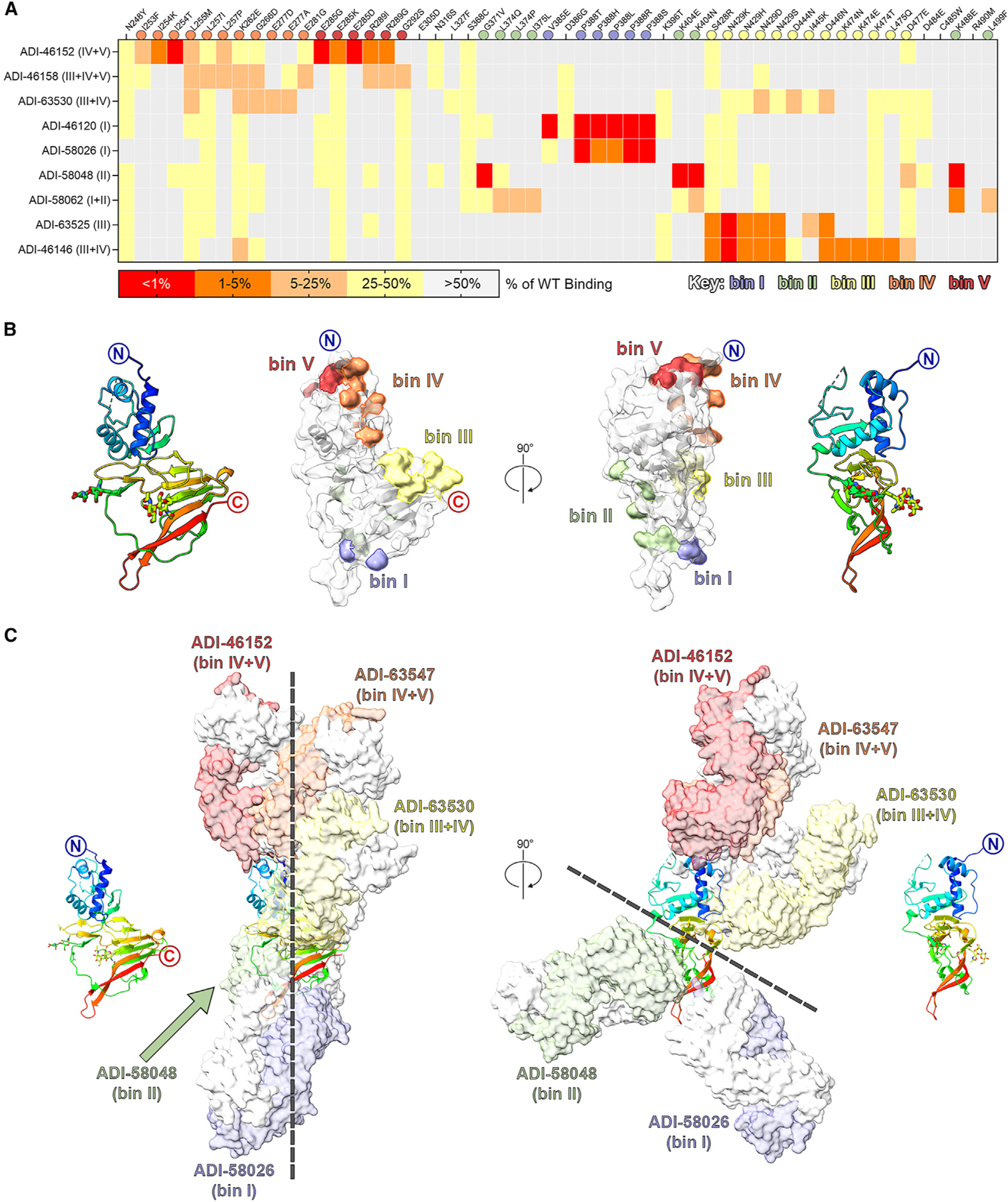
(A) Yeast-based mapping strategy of select antibodies to identify critical binding residues on GP38. The percentage of antibody binding retained by each GP38 variant is colored according to the key. Critical residues are defined as mutations that led to a binding disruption of 75% or more and are colored by the assigned antigenic site.
(B) Yeast-based critical residues mapped on the surface of GP38: bin I (blue, residues Val385, Pro388), bin II (green, residues Gly371, Leu374, Ile375, Lys404, Lys488, Leu499), bin III (yellow, residues Ser428-Ala429, Asp444-Asp446, Lys474-Leu475, Asp477), bin IV (orange, residues Ile253-Leu255, Leu257, Lys262, Gly266, Glu277, Glu281), bin V (red, residues Glu285, Arg289, Gly292).
(C) Composite structure of GP38 bound with representative antibodies. GP38 is shown as a rainbow ribbon and Fabs as molecular surfaces. Heavy chains are colored to represent the five non-overlapping bins, and light chains are white. Black dashed lines highlight the vertical alignment of Fabs along one plane (left) and the opposing binding directions to another plane (right).
