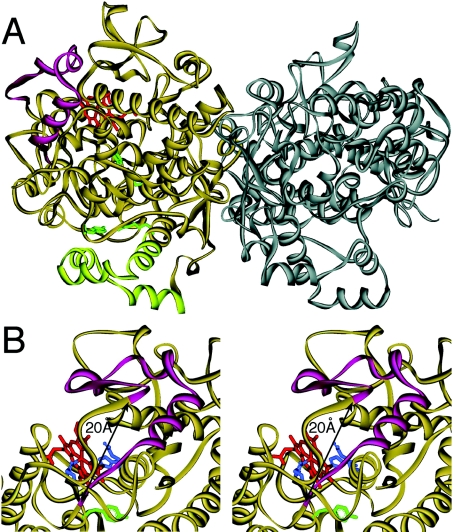
Figure 8. Three-dimensional structure of ovine COX-1 illustrating the location of amino acids Val-396 to Arg-432.
A ribbon diagram of the crystal structure of the ovine COX-1 homodimer is shown [22]. (A) The 37 amino acids from Val-396 to Arg-432 (magenta) are located on the far side of the dimer interface forming a loop near the haem binding (peroxidase) site. The four membrane binding helices at the bottom are shown in light green, four critical oxygenase active site residues (Arg-120, Tyr-355, Tyr-385 and Ser-530; ovine COX-1 numbering) are in green and the haem group in the peroxidase active site is depicted in red. (B) Stereo view of the haem binding site. Compared with (A), the protein is turned by approx. 30° to the right. The distance between the α-carbon atoms of Lys-395 and Ile-433 is approx. 20 Å. The distal (Gln-203 and His-207) and proximal (His-388) haem ligands are in blue and Tyr-385 is in green.