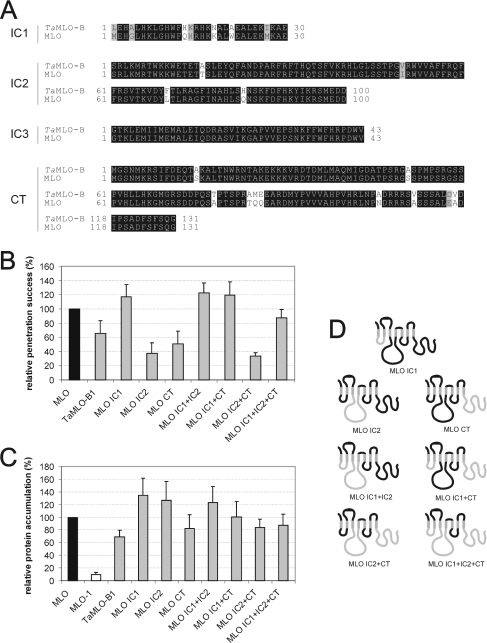
Figure 5. Domain-swap analysis with intracellular domains of barley MLO and TaMLO-B.
(A) Amino acid sequence alignment of the first to third intracellular loop (IC1–IC3), as well as the CT of MLO and TaMLO-B1. (B) Functional assay of single-amino-acid-substitution MLO variants. Leaf segments of the powdery-mildew-resistant barley cultivar BC Ingrid mlo-5 were co-bombarded with a GUS reporter construct and the bifunctional plasmid pUGLUM (encoding the GFP reporter plus wild-type MLO) or mutant versions thereof (encoding variants CT swap, CT swap A417S or CT swap A453S). Leaves were then inoculated with Bgh K1, and were stained for GUS activity and fungal structures as described in the Materials and methods section. (C) Assessment of MLO protein accumulation. Relative accumulation of wild-type MLO and mutant version MLO-1, as well as variants TaMLO-B1, MLO IC1, MLO IC2, MLO CT, MLO IC1+IC2, MLO IC1+CT, MLO IC2+CT and MLO IC1+IC2+CT was determined by dual-luciferase assays of transfected A. thaliana protoplasts as described in the Materials and methods section. Note that the values obtained for MLO and MLO-1 result from a common experiment with the constructs shown in Figures 2(C) and 2(E). (D) Schematic representation of the domain-swap constructs MLO IC1, MLO IC2, MLO CT, MLO IC1+IC2, MLO IC1+CT, MLO IC2+CT and MLO IC1+IC2+CT. Black bends illustrate MLO portions, whereas light-grey bends depict the respective TaMLO-B1 fraction.