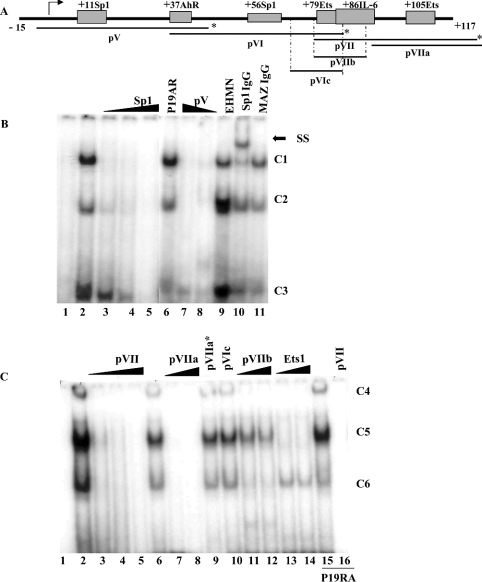
Figure 3. EMSA analysis of the human SMN core promoter.
(A) Schematic representation of the proximal human SMN promoter (nt −15 to +117) and the putative cis-elements under investigation. Indicated below are the double-strand DNA probes used for EMSA studies, pV, pVI and pVII as well as competitor probes pVIc, pVIIa and pVIIb. The asterisk marks the extremity that was end-labelled with [γ-32P]dCTP. (B) EMSA analysis using end-labelled pV probe (lane 1) incubated with nuclear extract prepared from P19 (lanes 2–5, 7, 8, 10 and 11), P19RA (lane 6) or EHMN (lane 9) cells. A double-strand, unlabelled competitor oligonucleotide corresponding to the consensus Sp1 sequence was added in 10, 50 or 200 times excess in lanes 3–5. Double-strand pV competitor was added in 100 or 50 times excess in lanes 7 and 8 respectively. Binding reactions in the presence of 2 μl of anti-Sp1 or anti-MAZ antibody were run in lanes 10 and 11 respectively. The three DNA–protein complexes are designated as C1, C2 and C3 as shown to the right of the gel. A single supershift (SS) complex was observed in lane 10 as indicated by the arrow. (C) EMSA analysis using end-labelled pVII probe (lane 1) incubated with 4 μg of nuclear extract prepared from P19 (lanes 2–14) or P19RA (lanes 15 and 16). Lanes 2, 6 and 15 do not contain competitor probes. Double-strand, unlabelled competitor probes designated at the top of the gel were added in 10× (lane 3), 50× (lanes 4, 7, 11 and 13) or 200× excess (lanes 5, 8–10, 12, 14 and 16). The three DNA–protein complexes are designated as C4, C5 and C6 as shown to the right of the gel. The bottom of the gel was removed (B, C) so that the band corresponding to free probe is not shown. EMSAs were repeated at least three times.