Figure 4.
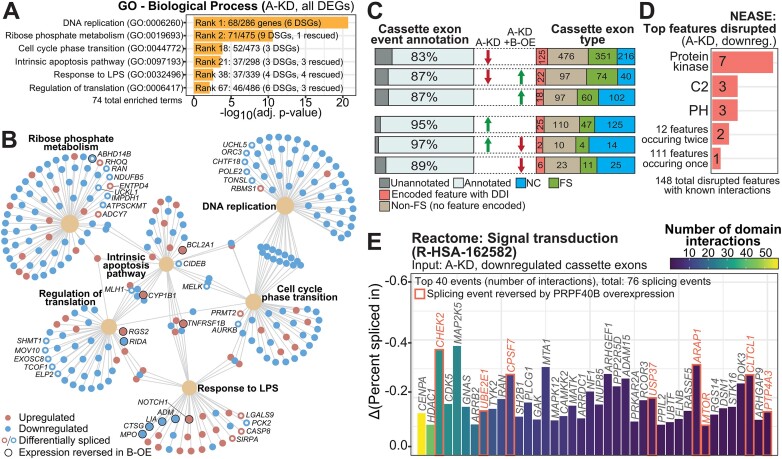
Enriched pathways and functions for the expression and splicing targets of PRPF40A. (A) Details of the Gene Ontology (GO) genesets selected for analysis of DEGs in the PRPF40A-KD (A-KD) dataset. DSGs: differentially spliced genes, DEGs that are also differentially spliced. The rescued genes refer to DEGs whose changes upon PRPF40A-KD were dampened by PRPF40B-OE. (B) Network plot of DEGs in the GO genesets indicated in Figure 4A. DEGs with concomitant differential splicing are indicated by hollow circles, while a black border indicates that the DEG was rescued by PRPF40B overexpression. (C) Distribution of cassette exon types by annotation for coding changes as in the key below, in addition to the percentage of differentially spliced cassette exons that are annotated as alternative exons in the GENCODE V43. (D) The three most numerous protein feature types suppressed by PRPF40A-KD, as reported by NEASE. PH: Pleckstrin homology domain. (E) The top 40 cassette exons with encoded protein features that are downregulated upon PRPF40A depletion, ranked by the number of known domain-domain interactions. Red borders indicate that the splicing event was rescuable by PRPF40B overexpression.