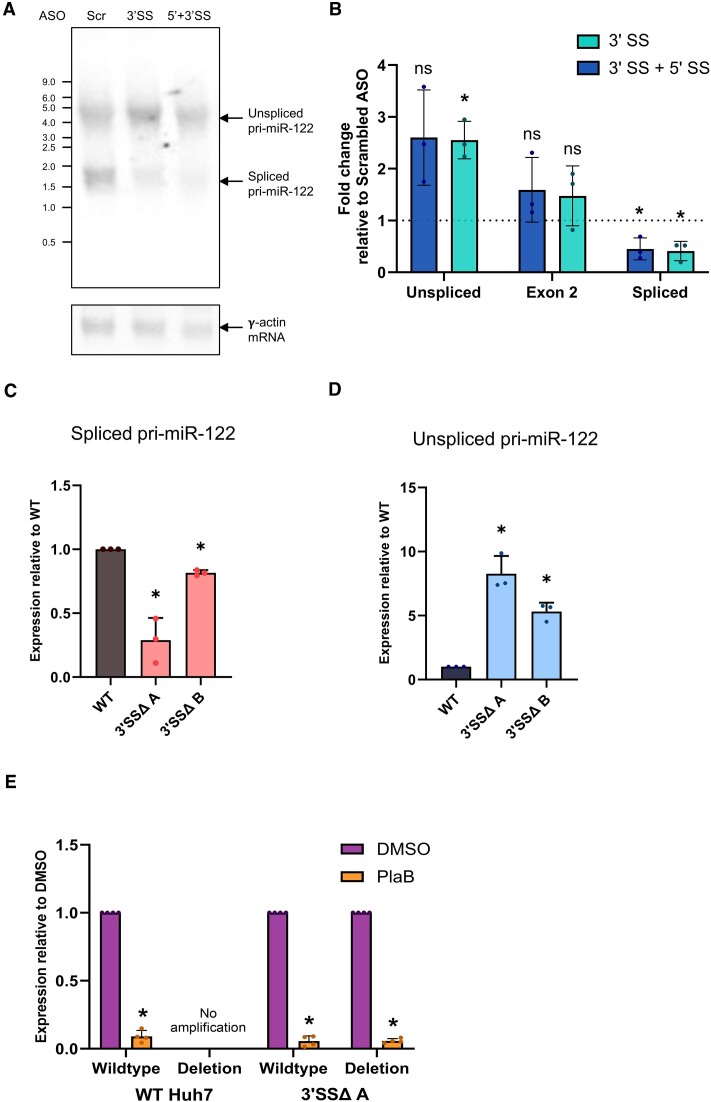
Figure 6.
Pri-miR-122 3′SS inhibition or mutation does not inhibit transcription. (A) Northern blot showing reduction of spliced, but not unspliced, pri-miR-122 in total RNA extracted from Huh7 cells following transfection of ASOs targeting the 3′SS, or 5′SS + 3′SS, compared to a scrambled control ASO. γ-Actin mRNA is shown as a loading control. (B) Chromatin-associated RNA was extracted from ASO-transfected cells and analysed by RT-qPCR with primer pairs specific to pri-miR-122, as shown in Figure 2A, relative to GAPDH pre-mRNA. (C) CRISPR/Cas9 modification was used to delete the region spanning the branch point, polypyrimidine tract and 3′SS of pri-miR-122 in Huh7 cells. Total RNA was extracted from two clonal cell lines heterozygous for the mutation (3′SSΔ A and 3′SSΔ B), and WT Huh7 cells cultured in parallel, and analysed by RT-qPCR with primers specific to spliced pri-miR-122. Pri-miR-122 was normalized to 18S rRNA and is shown relative to WT Huh7. (D) As panel (C), except that unspliced pri-miR-122 was analysed by RT-qPCR. (E) WT Huh7 cells or heterozygous 3′SSΔ clone A cells were treated with PlaB and total RNA extracted. RT-qPCR with primers specific to the WT or mutant (deletion) unspliced pri-miR-122 was used to determine the effects of PlaB relative to a DMSO-treated control, with data normalized to 18S rRNA. Northern blot is representative of two independent experiments and all other data represent mean of at least three independent experiments, with error bars showing SD. *P< 0.05; n.s., not significant.