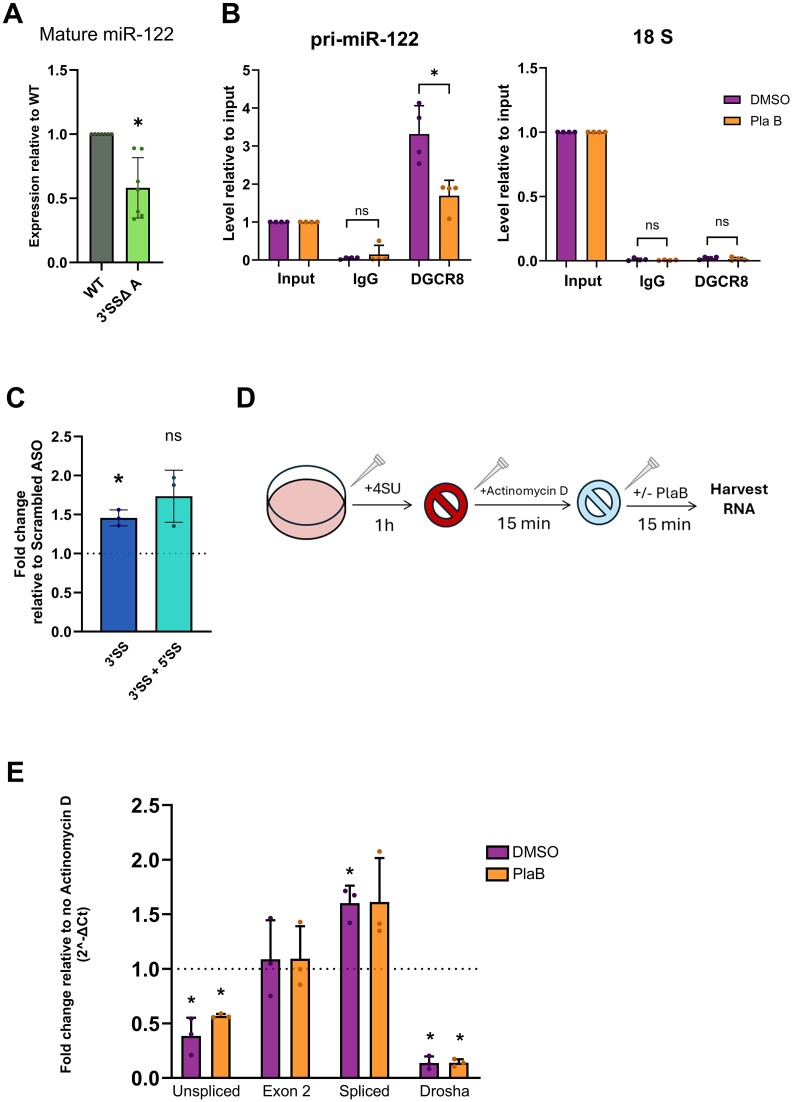
Figure 7.
Splicing promotes cotranscriptional, but not post-transcriptional, Microprocessor cleavage of pri-miR-122 independent of transcription. (A) Mature miR-122 levels in 3′SSΔ A were determined by RT-qPCR, normalized to U6 snRNA and shown relative to WT Huh7 cells cultured in parallel. (B) RIP was carried out using an antibody to endogenous DGCR8 in Huh7 cells ± PlaB. RT-qPCR with primers 5′ of the pre-miR-122 hairpin was used to measure pri-miR-122 in DGCR8 RIP, or an IgG control, relative to 10% input. 18S rRNA was measured by RT-qPCR as a control for non-specific binding. (C) Chromatin-associated RNA was extracted from ASO-transfected cells and analysed by RT-qPCR with primers spanning the Drosha cleavage site on pri-miR-122, relative to GAPDH pre-mRNA. (D) Diagram showing the approach used to investigate pri-miR-122 processing following transcription inhibition in panel (E). (E) 4SU-labelled RNA was isolated from Huh7 cells ± PlaB treatment following Act D treatment, and analysed by RT-qPCR using primer pairs shown in Figure 2A. RT-qPCR with different sets of primers for pri-miR-122 was used to detect changes in 4SU-labelled RNA from DMSO- and PlaB-treated cells relative to a no Act D control, normalized to 18S rRNA. Data represent mean of at least three independent experiments, with error bars showing SD. *P< 0.05; n.s., not significant.