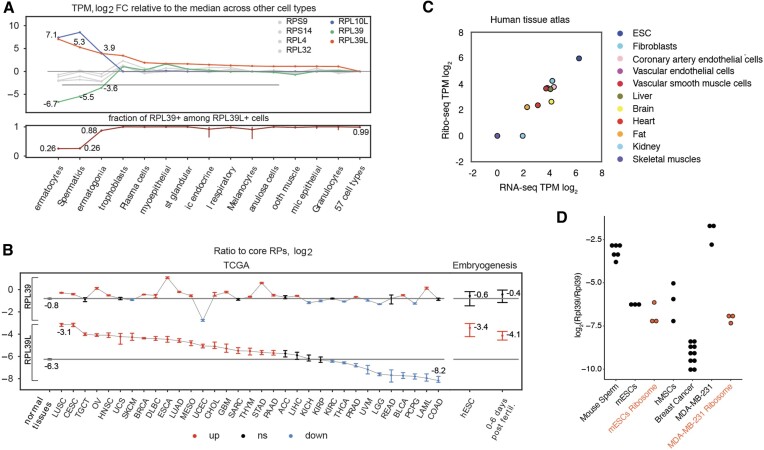
Figure 1.
RPL39L expression across cell types. (A) Top: HPA-provided normalized gene expression values (transcript-per-million, TPM) were used to identify the 14 cell types with highest RPL39L expression. The log2 fold change in each of these cell types relative to the median across all other 57 cell types in HPA is shown for RPL39L (orange), RPL39 (green), RPL10L (blue) genes and 4 core RPs (gray). Bottom: the proportion of RPL39L+ cells that also contained RPL39-derived reads in single cells of the types shown in the top panel. (B) Ratio of RPL39 and RPL39L to core RP expression (log2) in bulk RNA-seq samples of primary tumors from TCGA (https://www.cancer.gov/tcga/) (left panel) as well as human pre-implantation embryos and cultured embryonic stem cells (17) (right panel). See https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations for TCGA cancer-type abbreviations. Black horizontal lines show the median values of these ratios in adult normal tissue samples from TCGA. Statistically significant (two-sided Wilcoxon test, Benjamini–Hochberg FDR < 0.05) positive and negative deviations from the medians are shown in orange and blue, respectively. For each category, the 95% confidence interval over all samples (for bulk data from TCGA) or all cells (for single-cell data) is shown. Categories for which the ratios were not significantly different from the median of the normal samples are shown in black. (C) Ribo-seq vs. RNA-seq level expression of RPL39L in samples from the human tissue atlas (extracted from the supplementary material of (63)). (D) Quantification of RPL39L/RPL39 protein ratio in various cellular systems (mouse sperm cells −6 samples, breast cancer cell line MDA-MB-231, bone marrow-derived mesenchymal stem cells (hMSC) and E14 mouse embryonic stem cells (mESC) −3 independent samples, breast cancer tissues −10 samples) using reference peptides. Similar quantification from purified ribosomes of E14 and MDA-MB-231 cells is also shown.