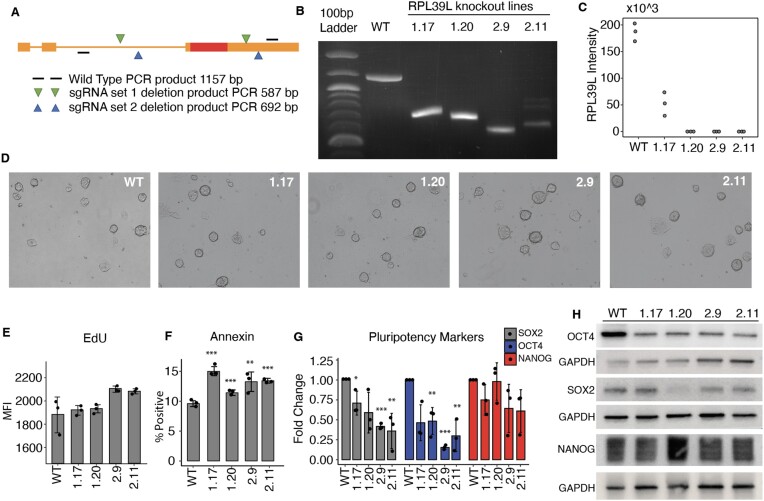
Figure 2.
Characterization of RPL39L KO mESCs. (A) Schema of sgRNA design. Two distinct pairs of sgRNAs (in green and blue) were designed to target flanking regions of the RPL39L coding sequence (CDS, shown as a red box). Primers (black lines) from further upstream and downstream in the RPL39L locus were used for amplification, and the expected sizes of the PCR products are indicated for both the WT locus (1157 nts) and the edited loci (587 and 692 nts, respectively, for the sgRNA sets 1 and 2). (B) PCR products from the WT E14 cells and the 4 independent clones, 1.17 and 1.20 generated with sgRNA set #1 and2.9 and 2.11 generated with sgRNA set #2. (C) Expression of RPL39L in individual clones determined with targeted proteomics (n = 3 for each clone). (D) Representative bright field images of colonies from all analyzed clones. (E) Results of the ethynyldeoxyuridine (EdU, thymidine analog) incorporation assay. EdU-treated cells were fixed, permeabilized and the AF488 fluorophore was linked to the EdU in the replicated DNA by click-chemistry. The label intensity was measured by FACS (Mean fluorescence intensity, MFI). (F) Results of the annexin binding assay. Cells were incubated with AF488-conjugated Annexin V and counterstained with phycoerythrin (PE). The proportion of AF488+PE− (apoptotic cells) relative to the parent cell population was determined by FACS. (G) RT-qPCR of the pluripotency factors SOX2, OCT4 and NANOG. Values are 2−ΔΔCt, relative to RRM2 (internal reference) and to WT. (H) Representative Western blots of pluripotency markers SOX2, OCT4 and NANOG in the RPL39L KO lines and WT relative to GAPDH. In panels F and G, *, ** and *** correspond to P-values <0.05, <0.01 and < 0.001, respectively in the two tailed t-test comparing KO lines with WT.