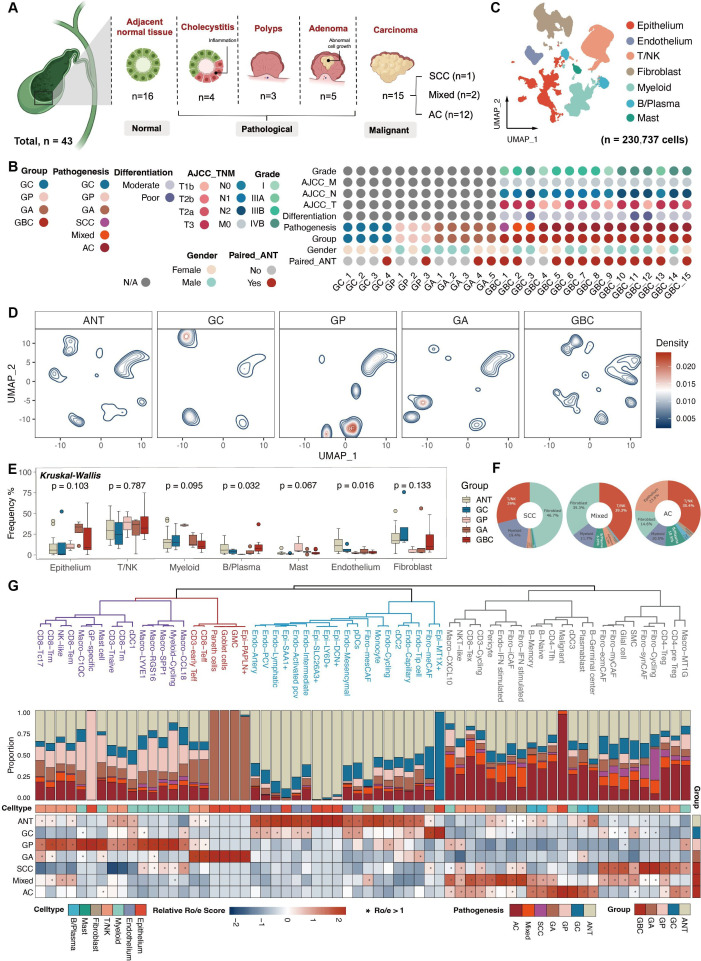
Figure 1.
Landscape of precancerous lesions of gallbladder and gallbladder cancer by scRNA-seq of 43 samples. (A) Overview of the study design and sample composition. (B) Clinicopathological profiles of samples enrolled in scRNA-seq cohort. (C) Uniform Manifold Approximation and Projection unveiling seven major cell lineages. (D) Visualisation of alterations in major lineages composition among groups through cell density mapping. (E) Boxplots revealing the frequency of major cell compartments across different groups. Statistical significance was evaluated via the Kruskal-Wallis’ test. (F) Pie charts illustrating the predominant lineage composition among diverse GBC subtypes. (G) Phenotypic relationships and population abundance of 64 cell subsets excluding five patient-specific clusters. Unsupervised hierarchical clustering of cell subsets (top panel). Bar plot showing the distribution of cell subsets across seven tissue subtypes (middle panel). Heatmap at the bottom showing tissue prevalence estimated by Ro/e score for each cell subset. AC, adenocarcinomas; AJCC, American Joint Committee on Cancer; ANT, adjacent normal tissues; GA, gallbladder adenomas; GBC, gallbladder cancer; GC, gallbladder cholecystitis; GP, gallbladder polyps; NK, natural killer; SCC, squamous cell carcinoma; sc-RNA-seq, single-cell RNA sequencing; TNM, tumour, node, metastasis.