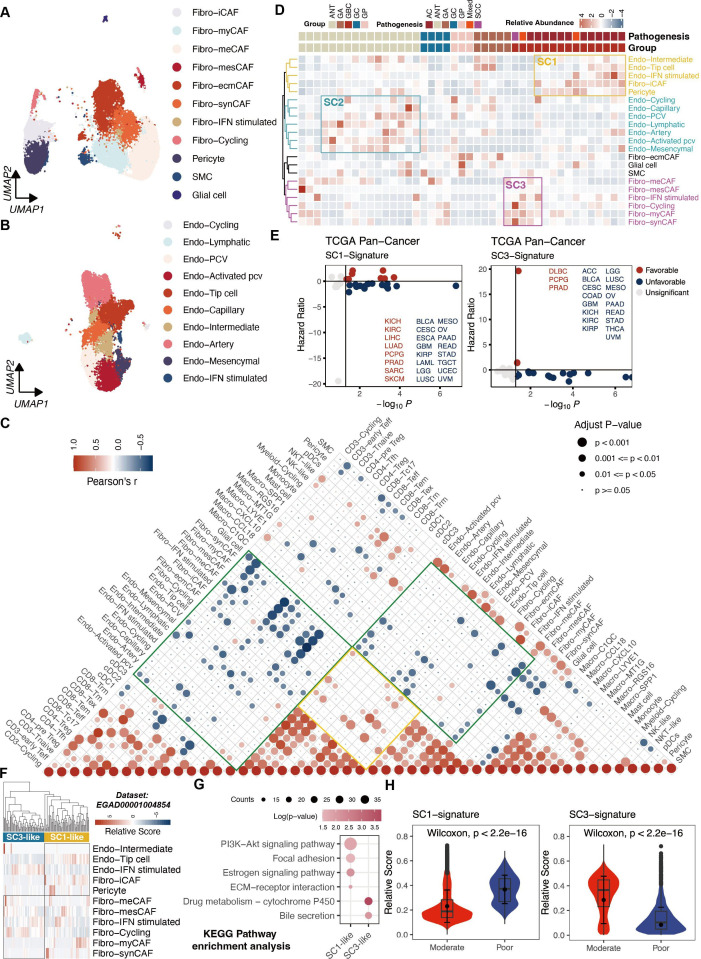
Figure 3.
Phenotypic abundance and interactions of stromal cells. (A) UMAP of subpopulations of fibroblasts. (B) UMAP of subpopulations of endothelial cells. (C) Correlation analysis among TME subsets in all samples based on corresponding relative abundance. P values were calculated using the Spearman correlation test with Benjamini-Hochberg correction for multiple comparisons. (D) Inference of three ecotypes based on stromal cell compositions in the 43 samples. (E) Survival analysis of SC1 and SC3 gene signature across all cancer types from TCGA. Optimal cut points were determined using the survminer package. (F) Deconvolution of SC1 and SC3 cellular ecotypes in cohort by Pandey et al. (G) KEGG enrichment analysis of SC1 and SC3 ecotypes. (H) Violin plots showing significant differences in signature scores of SC1 and SC3 between patients with well-differentiated and poor-differentiated GBC. AC, adenocarcinomas; ANT, adjacent normal tissues; ECM, extracellular matrix; GA, gallbladder adenomas; GBC, gallbladder cancer; GC, gallbladder cholecystitis; GP, gallbladder polyps; KEGG, Kyoto Encyclopedia of Genes and Genomes; SCC, squamous cell carcinoma; TCGA,The Cancer Genome Atlas; TME, tumour microenvironment; UMAP, Uniform Manifold Approximation and Projection.