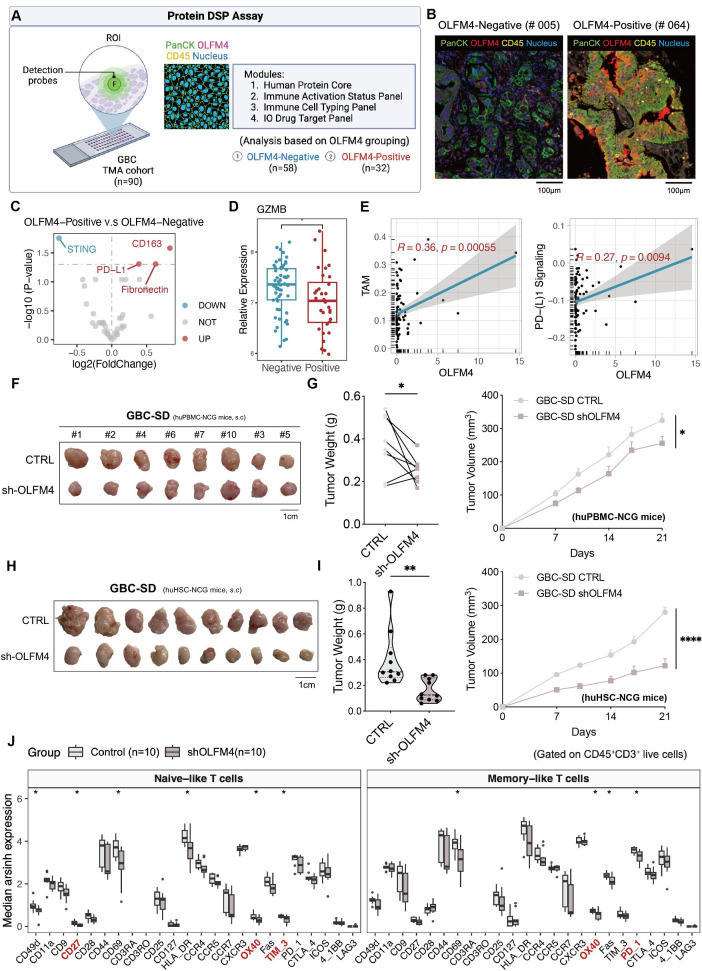
Figure 6.
OLFM4 positively correlated with TAM infiltration and suppressed antitumour T-cell immunity in vivo. (A) Overview of DSP assay design to investigate spatial heterogeneity of GBC. (B) Representative images of the OLFM4-negative sample and OLFM4-positive sample. (C) Volcanic plot of differentially expressed proteins among groups stratified by OLFM4 expression. (D) Box plot comparing GZMB expression between groups. (E) Pearson correlation of TAM and PD-(L)1 signalling with OLFM4 expression. (F) GBC-SD CTRL/sh-OLFM4 cells were injected subcutaneously into the right flank of NCG mice and transferred with activated PBMC to build the xenograft model. Gross morphology of tumours in the huPBMC-NCG model (CTRL, n=8; sh-OLFM4, n=8). (G) Tumour volume (left) and tumour weight (right) of F were calculated. (H) Subcutaneous injection of GBC-SD CTRL/sh-OLFM4 cells into CD34+ humanised mice to obtain tumour xenografts. Gross morphology of tumours in the huHSC-NCG model (CTRL, n=10; sh-OLFM4, n=10). (I) Tumour volume (left) and tumour weight (right) of H were calculated. (J) Relative expression levels of functional markers of naive-like+ T cells and memory-like+ T cells with tumours in H were determined by cytometry by time of flight. *p<0.05 using a Wilcoxon test. CyTOF, mass cytometry or cytometry by time of flight; DSP, digital spatial profiler; GBC, gallbladder cancer; GZMB, granzyme B; OLFM4, olfactomedin 4; PBMC, peripheral blood mononuclear cells; PD-L1, programmed cell death ligand 1; ROI, region of interest; TAM, tumour-associated macrophage.