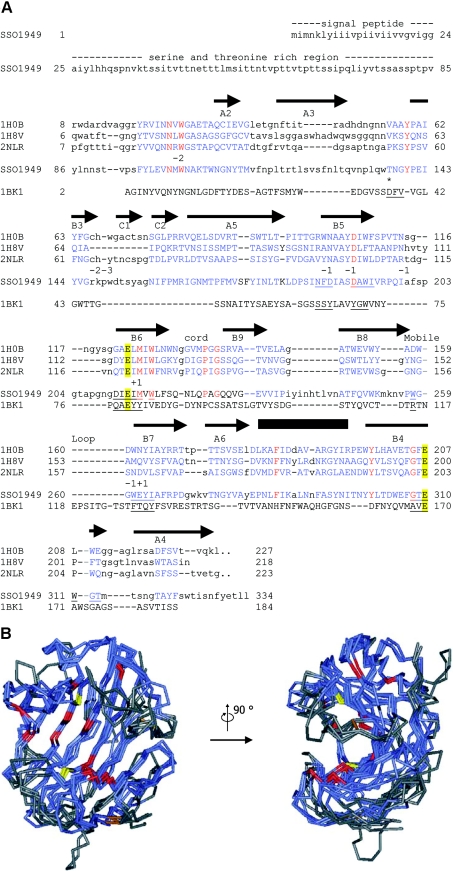
Figure 1. Sequence alignment and structural model of SSO1949.
(A) Aligned are the catalytic modules of the cellulases from Rhodothermus marinus (Protein Data Bank no. 1H0B) [9], T. reesei (PDB no. 1H8V) [8], Streptomyces lividans (PDB no. 2NLR) [7] and S. solfataricus SSO1949. The complete sequence is given only for SSO1949. The first 24 amino acids are predicted to be a signal sequence followed by a serine- and threonine-rich region (amino acids 25–85), which has no sequence similarity to other proteins. Regions of the GH domains which could be aligned are shown in blue capital letters. Identical residues are in red. The catalytic glutamate residues Glu-213 and Glu-310 are emphasized with yellow boxes. The arrows (β-strands) and the box (α-helix) above the sequence show the positions of the secondary-structure elements of the R. marinus cellulase. The elements are labelled according to Crennell et al. [9]. The numbers below the sequence 2NLR indicate the subsites of the active-site cleft. The alignment also includes the extremely acidophilic xylanase from A. kawachii (PDB no. 1BK1) [24]. This enzyme belongs to the GH family 11, which has a fold similar to the GH family 12 [4]. For the acidophilic enzymes from S. solfataricus and Aspergillus kawachii, residues within 0.5 nm of the catalytic glutamate residues are underlined. Thr-137 is homologous with Asp-37 of the A. kawachii xylanase, which is implicated in the low-pH optimum of this enzyme (*, see text for details). The alignment of the two acidophilic enzymes along with the neutral cellulases does not reveal any particular amino acid which could be responsible for the low pH optimum of the S. solfataricus enzyme. (B) The structure of SSO1949 was modelled with ESyPred3D [21] using the cellulase from R. marinus (PDB no. 1H0B) as template. Shown are the backbones of the model structure (amino acids 86–329) superimposed with the structures of the cellulases from R. marinus, T. reseii (PDB no. 1H8V) and S. lividans (PDB no. 2NLR). In blue are the parts of the protein which could be structurally aligned. Structural alignment was not possible for the regions in grey. The backbone trace is red for amino acids which are completely conserved within the four proteins. The catalytic glutamate residues are depicted in yellow. Disulphide bridges are displayed in brown. The region of the active site seems to be structurally well conserved and contains most of the completely conserved amino acids. The active-site cleft is best viewed from the side (right picture).