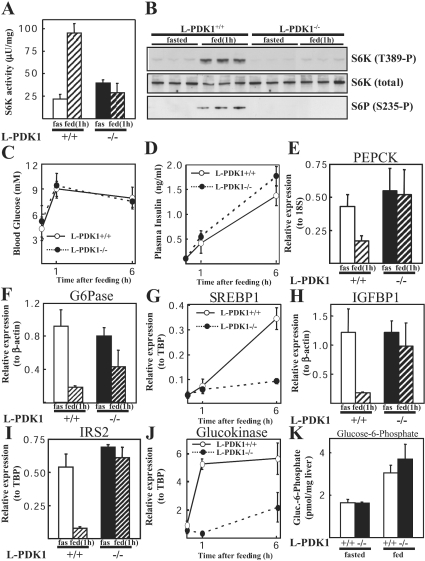
Figure 5. Regulation of gene expression by feeding.
The indicated mice were fasted overnight and then allowed to re-feed ad libitum for 1 or 6 h. Blood was taken, and the livers were then rapidly extracted and frozen in liquid nitrogen. (A) S6K1 was immunoprecipitated from liver extracts, and the activity was determined using a quantitative peptide phosphorylation assay. Each point represents the mean activity±S.D. of three different livers with each assayed in triplicate. fas, fasted. (B) Extracts were also immunoblotted with the indicated antibodies. S6P, S6 ribosomal protein. (C) Blood glucose and (D) plasma insulin levels were measured. The data are shown as the mean±S.D. for triplicate mice for glucose and duplicate mice for insulin. (E)–(I) mRNA levels for specific genes were quantified in livers of the indicated mice as described in the Materials and methods section. RPA analysis was performed for expression of G6Pase (F) and IGFBP1 (H). Real-time PCR analysis was performed for expression of PEPCK (E), SREBP1 (G), IRS2 (I) and glucokinase (J). Three mice were analysed in triplicate for each data point, except for the 1 h time point glucokinase data in (J) that was analysed in duplicate. Data is presented as relative levels (±S.E.M.) of G6Pase or IGFBP1 mRNA to β-actin control, IRS2, SREBP1 and glucokinase to TBP control, or PEPCK to 18 S control. Unless stated, animals were re-fed for 1 h before RNA extraction. (K) Hepatic glucose 6-phosphate levels were measured in liver extracts derived from mice fed ad libitum and from mice that had been fasted overnight (16 h). The results are expressed as the means±S.D. from three animals for each condition.