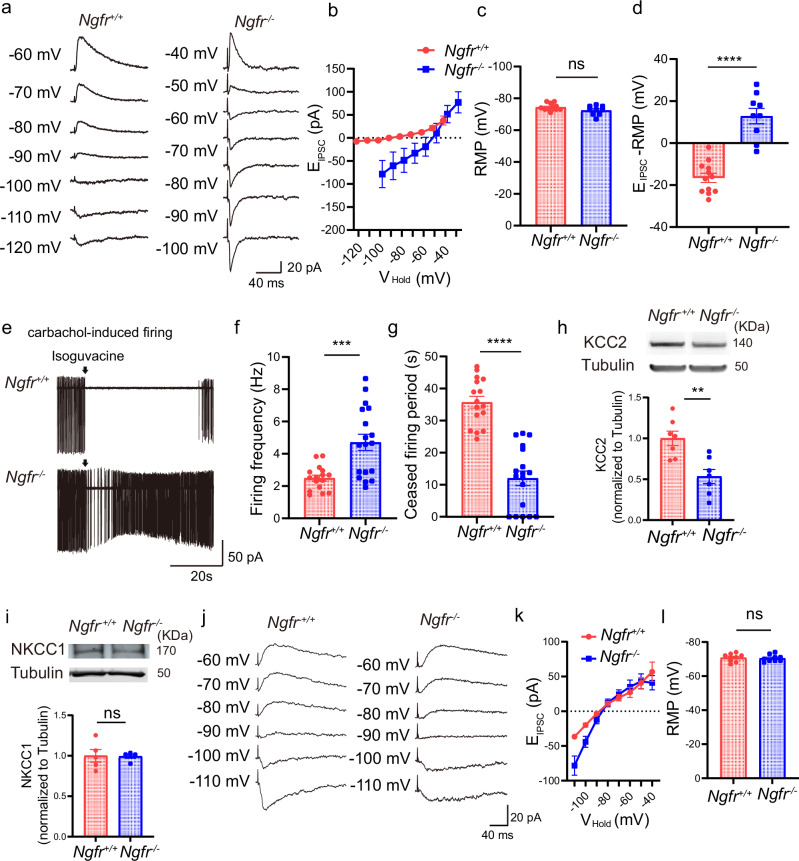
Fig. 6. Depolarizing-shifted reversal potential of GABAA-mediated EIPSCs and reduced GABAergic inhibition in Ngfr−/− mPFC.
a Perforated patch recorded the reversal potential of EIPSCs and resting membrane potential (RMP) in the layer V pyramidal neurons, in the presence of 50 µM APV and 50 µM CNQX. Representative traces indicate a depolarizing shift of EIPSCs in Ngfr−/− mPFC. b Group summary suggests depolarizing shift of reversal potential of EIPSCs in Ngfr−/− mice. Ngfr+/+, n = 12 cells from 5 mice; Ngfr−/−, n = 9 cells from 5 mice. c No differences of RMP in current-clamp mode between genotypes. p = 0.095. Ngfr+/+, n = 12 cells from 5 mice; Ngfr−/−, n = 9 cells from 5 mice. d Hyperpolarizing GABAergic driving force in Ngfr+/+ mice, versus depolarizing-shifted GABAergic strength in Ngfr−/− mice. p < 0.0001. Ngfr+/+, n = 12 cells from 5 mice; Ngfr−/−, n = 9 cells from 5 mice. e Reduced GABAergic inhibition of layer V pyramidal neurons in Ngfr−/− mPFC. Cell-attached recordings of carbachol-induced firing. Representative traces of basal spike frequency and effects of puffing GABA agonist isoguvacine (100 µM). f In steady-state, carbachol-induced firing frequency was significantly higher in Ngfr−/− mice. p = 0.0004. Ngfr+/+, n = 16 cells from 4 mice; Ngfr−/−, n = 18 cells from 4 mice. g After puffing isoguvacine, ceased firing period was significantly shorter in Ngfr−/− mice. p < 0.0001. Ngfr+/+, n = 16 cells from 4 mice; Ngfr−/−, n = 18 cells from 4 mice. h Representative blots and quantification of KCC2 levels in mPFC. Significantly reduced KCC2 expression in Ngfr−/− mice. p = 0.0026. Ngfr+/+, n = 7; Ngfr−/−, n = 7. i Representative blots and quantification indicate intact NKCC1 expression in Ngfr−/− mPFC. p = 0.93. Ngfr+/+, n = 5; Ngfr−/−, n = 5. j Perforated patch recorded the reversal potential of EIPSCs and RMP of hippocampal CA1 pyramidal neurons. k Intact reversal potential of EIPSCs in CA1 pyramidal neurons of Ngfr−/− mice. Ngfr+/+, n = 9 cells from 3 mice; Ngfr−/−, n = 9 cells from 3 mice. l No difference of RMP between genotypes. p = 0.75. Ngfr+/+, n = 9 cells from 3 mice; Ngfr−/−, n = 9 cells from 3 mice. Numerical data are means ± SEM. Statistical analyses were performed by unpaired two-sided t-test in (c), (d), (f–i), and (l). ***p < 0.001; ****p < 0.0001; ns not significant. Source data are provided as a Source Data file.