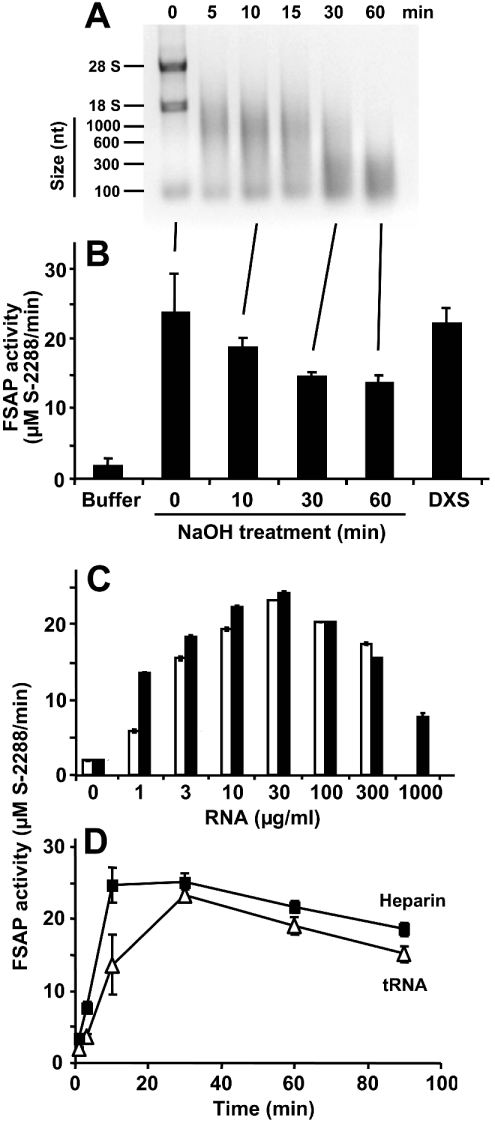
Figure 6. FSAP activation by an RNA-dependent template mechanism.
RNA was chemically hydrolysed by NaOH, and aliquots of the reaction mixture were withdrawn at the indicated time points and analysed by 1% agarose gel electrophoresis (A), followed by staining with ethidium bromide. RNA size markers, as well as 28 S and 18 S RNA, are indicated at the left margin. (B) In parallel, aliquots of the reaction mixture were analysed for FSAP cofactor activity as described. For comparison, FSAP activation was carried out by dextran sulphate (DXS). The results are expressed as the means±S.D. (n=3), and a representative experiment out of three is shown. (C) The (auto-)activation of single-chain FSAP (18.75 nM) was followed in the absence (0) or presence of the indicated concentrations of CHO cell total RNA (open bars) or yeast total RNA (closed bars), as described in the Experimental section. Note the bell-shaped concentration dependence, characteristic of a template mechanism. (D) The activation of single-chain FSAP (18.75 nM) by 100 μg/ml tRNA (△) or 5 units/ml heparin (■) in the presence of 1 mM CaCl2 was followed over time by a chromogenic substrate assay using S-2288. Samples were taken at the indicated time points and the values (corrected for FSAP activity in the absence of additives) are expressed as the means±S.D. (n=3), and a representative experiment out of three is shown.