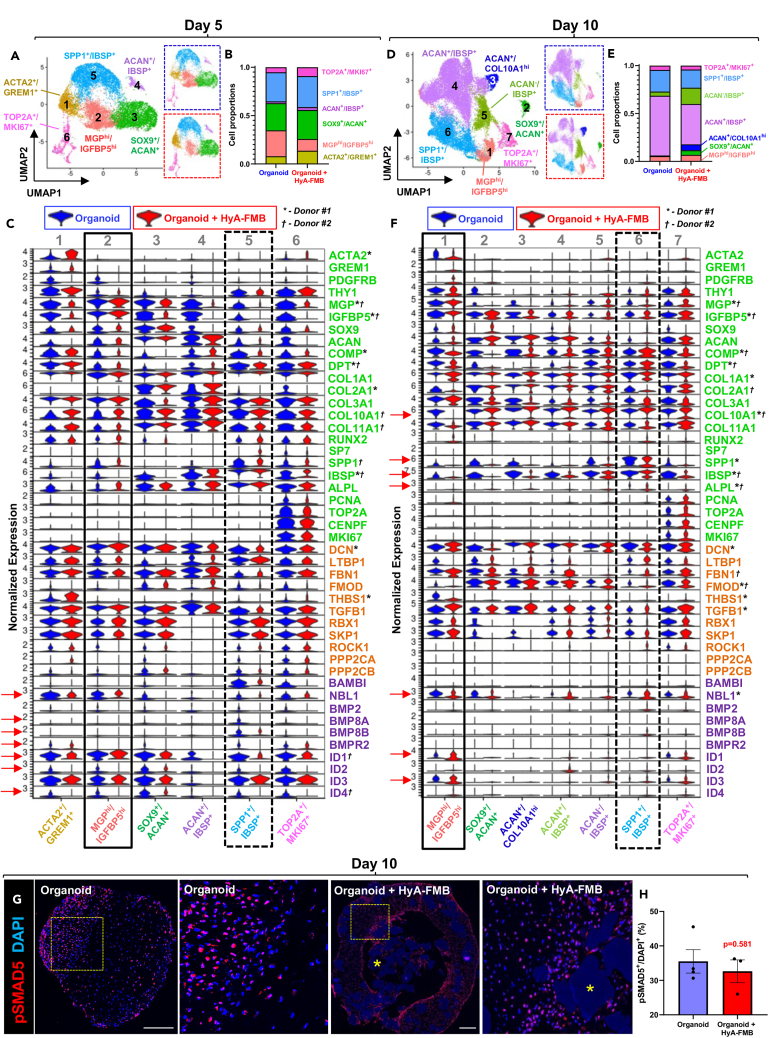
Figure 3.
HyA-FMBs restore BMP signaling in MGP/IGFBP5-enriched chondrogenic cells
(A–F) Cluster analyses from scRNA-seq of control (blue) and HyA-FMB (red) organoids at day 5 (A–C) and day 10 (D–F) of chondrogenic differentiation. Control and HyA-FMB datasets at each time point were normalized and integrated using Seurat. Marker genes for each cluster are listed in the corresponding color. Data are shown from donor #1.
(A, D) UMAP representation of integrated control and HyA-FMB datasets with annotated clusters. Split UMAPs are shown on the right for control organoids (blue dashed outline) and HyA-FMB organoids (red dashed outline).
(B and E) Bar chart depicting proportion of cell clusters in control and HyA-FMB datasets.
(C and F) Violin plots depicting gene expression split by experimental condition (blue violin plots are control organoids, red violin plots are HyA-FMB organoids) from integrated datasets at day 5 (C) and day 10 (F) of chondrogenic differentiation. Genes associated with TGF-β signaling (orange), BMP signaling (purple), and chondro-osteogenesis (green) are shown. Arrows depict notable gene expression differences (logfc>0.25, p < 0.05) between MGPhi/IGFBP5hi cluster (solid black box) and SPP1+/IBSP+ cluster (dashed black box). Global significance (logfc>0.25, p < 0.05) between control and HyA-FMB organoids is shown with an asterisk (∗) for donor #1 and dagger (†) for donor #2.
(G) Immunofluorescence of phospho-Smad5 (pSMAD5) in day 10 organoids. High-magnification insets shown to the right of their corresponding images. Nuclei counterstained with DAPI. Scale bars, 300 μm. Asterisks (∗) indicate HyA-FMBs.
(H) Quantification of pSMAD5+ nuclei in day 10 organoids. Each dot represents a biological replicate. Data are mean ± SEM (see also Figure S2).