Figure 3.
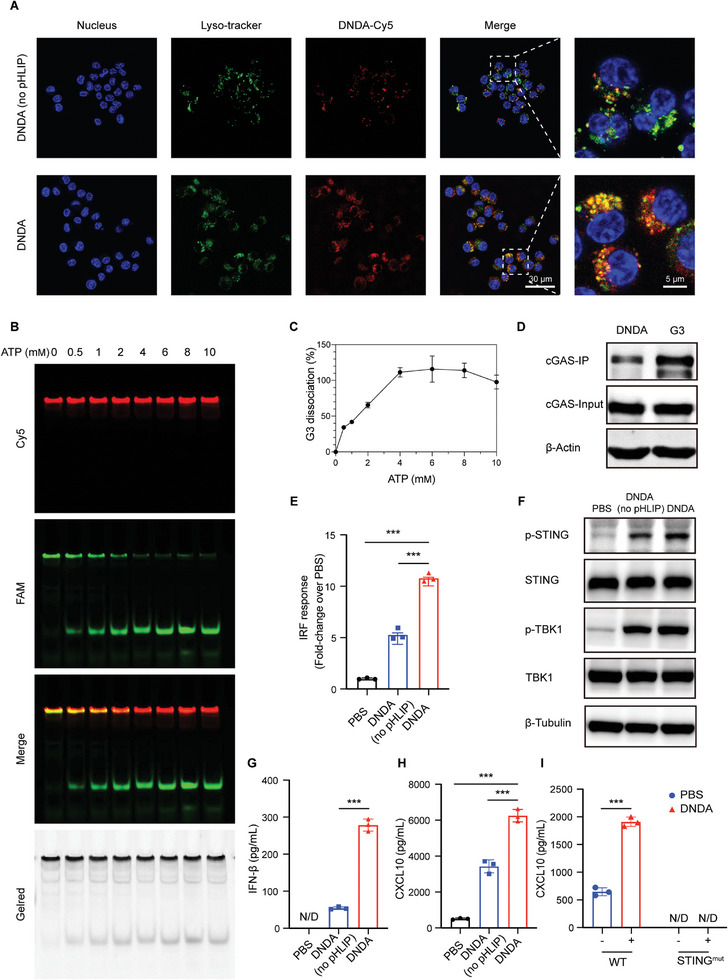
Programmable cGAS‐STING activation process of DNDA. A) Endosomal escape of Cy5‐labeled DNDA with or without pHLIP modification in RAW264.7 cells imaged by LSCM. (Scale bar: 30 µm) B) Fluorescent native PAGE analysis of dual fluorescent‐labeled DNDA treated with indicated concentration of ATP at 37 °C for 2 h. C) Quantitative grayscale analysis of G3 dissociation rate over ATP concentration. D) The binding between G3‐biotin, DNDA‐biotin, and cGAS using co‐IP analysis after co‐incubation with RAW264.7 lysate for 6 h. E) IFN regulatory factor (IRF) response elicited by DNDA with or without pHLIP in RAW ISG cells with an IRF‐inducible luciferase reporter. Data are shown as Mean ± SD (n = 3), and statistical significance was calculated via one‐way ANOVA with Tukey's post hoc test, *** p < 0.001. F) Immunoblotting assay of phosphate STING and TBK1 level in RAW264.7 cells after incubation with DNDA with or without pHLIP. G) IFN‐β and (H) CXCL10 expression level in RAW264.7 macrophages incubated with PBS and DNDA with or without pHLIP, as measured by ELISA assay. (N/D, not detected) Data are shown as Mean ± SD (n = 3), and statistical significance was calculated via one‐way ANOVA with Tukey's post hoc test, *** p < 0.001. I) CXCL10 expression level in DNDA‐treated BMDM cells from WT and STINGmut mice. (N/D, not detected) Data are shown as Mean ± SD (n = 3), and statistical significance was calculated via two‐way ANOVA with Sidak's post hoc test, *** p < 0.001.
