Figure 2.
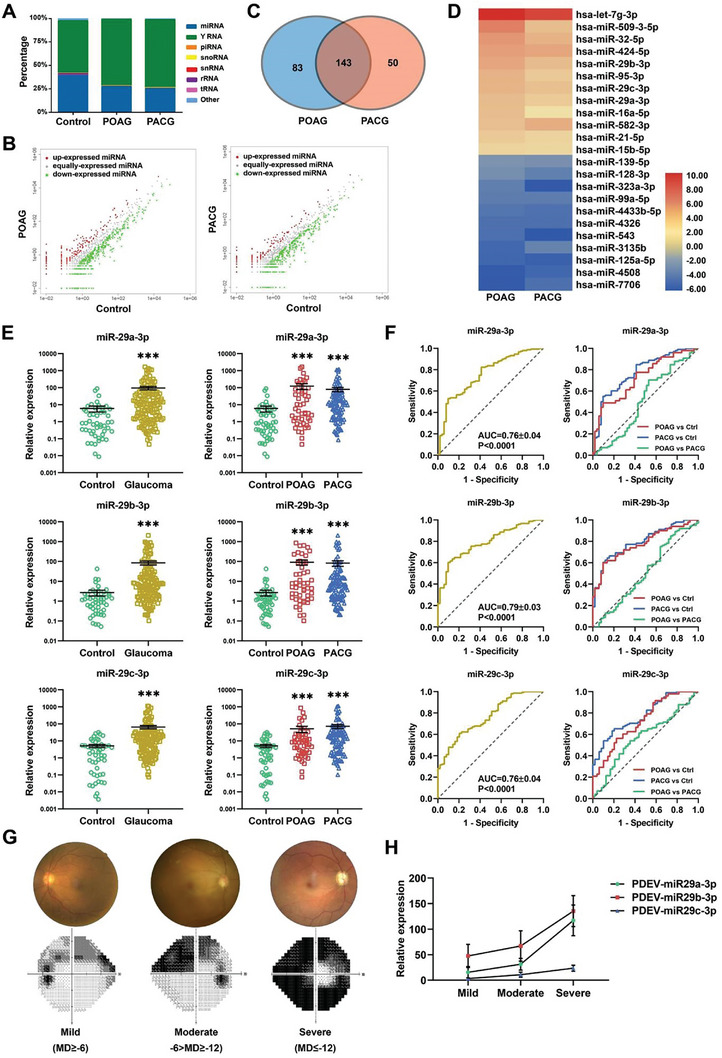
Identification and validation of candidate PDEV‐miRNAs with glaucoma specificity. A) Distribution of small RNAs detected by RNA‐seq in PDEV (n = 20 for each group; mt‐tRNA, transfer RNA located in the mitochondrial genome; rRNA, ribosomal RNA; piRNA, PIWI‐interacting RNA; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA). B) Scatter plot showing differentially expressed PDEV‐miRNAs in glaucoma patients compared to controls (fold change = 2 as the threshold). C) A Venn diagram showing that 143 differentially expressed PDEV‐miRNAs overlapped between POAG and PACG patients. D) Twenty‐three candidate miRNAs were selected from the RNA‐seq data, including the top 12 upregulated miRNAs and the top 11 downregulated miRNAs. E) qPCR analysis of individual patient samples confirmed that miR‐29s were significantly upregulated in both POAG and PACG patients (n = 51 for control and POAG, n = 102 for PACG). F) ROC analysis showing the diagnostic performance of PDEV‐miR‐29a‐3p, miR‐29b‐3p, and miR‐29c‐3p in POAG and PACG. G,H) Glaucoma patient classification based on MD values and PDEV‐miR‐29s expression increased with disease severity. (***p < 0.001).
