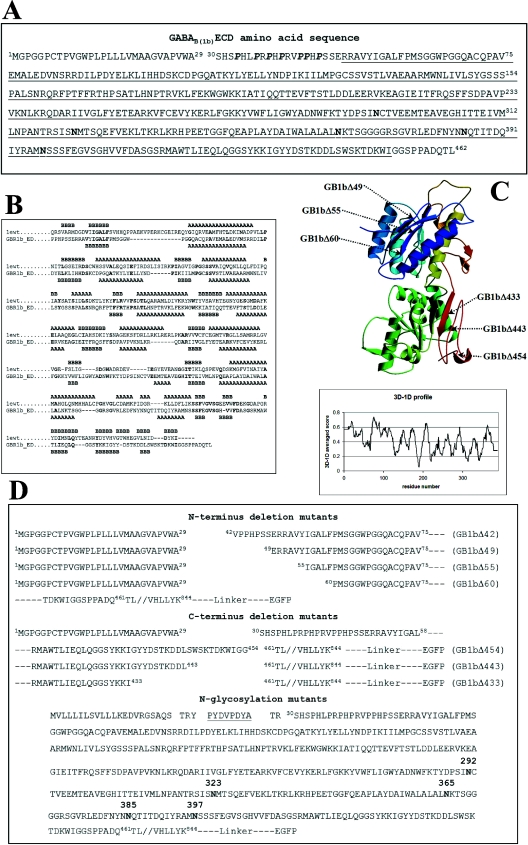
Figure 1. (A) GABAB(1b)ECD sequence, (B) sequence alignment, (C) model of the GABAB(1)ECD and (D) designed constructs.
The natural signal sequence of GABAB(1b) from R. norvegicus spans amino acids 1–29. Proline residues belonging to the N-terminal domain characteristic for the variant 1b appear in bold italic. The sequence encoded by exons 6–14 is underlined. The five potential N-glycosylation sites are indicated in boldface. (B) The amino acid sequence of the mGluR1 (1ewt) is derived from the PDB number 1EWT; GBR1b_ED is the sequence of the GABAB(1b)ECD. Secondary structural elements observed (in the crystal structure 1EWT) or predicted (for GABAB(1)ECD) are indicated with A for α-helices and B for β-strands. Identical residues in the two sequences appear in boldface. The following regions were omitted from the alignment used to generate the model: in mGluR1, residues 132–153 (not present in the PDB number 1EWT) and 348–405; in the GABAB(1)ECD sequence, residues 316–347. (C) The model was ramp-coloured from blue to green and yellow to red, going from the N- to the C-terminus. Arrows indicate the generated GABAB(1b) receptor deletion mutant proteins. The panel shows the plot obtained from the Verify3D algorithm using the co-ordinates of the modelled GABAB(1)ECD. The x-axis indicates the residue numbers and y-axis gives the averaged 3D–1D scores. (D) The top part of the panel shows the N-terminal deletion mutants, all containing the natural signal sequence of the GABAB(1b); the middle part of the panel shows the C-terminal deletions of the ECD. Details on construct generation are given in the Materials and methods section. The bottom part of the panel shows the sequence of the N-glycosylation mutants. The haemagglutinin tag is underlined. Asparagine residues, which can potentially be glycosylated in vivo, appear in boldface. All constructs are fused C-terminally through a short linker to the EGFP.