Fig. 1: Context independent on target chromatin access of Myt1l.
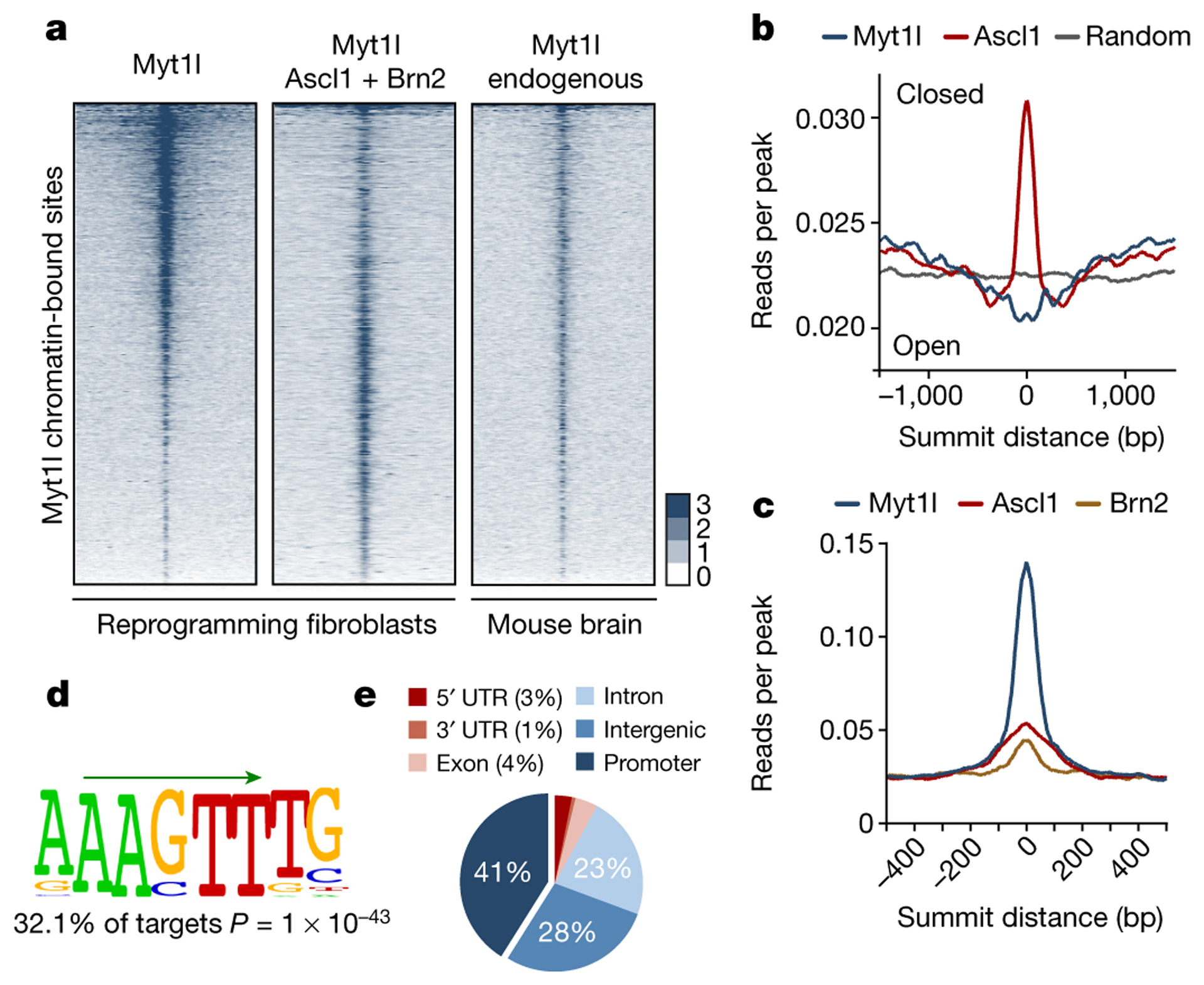
a, Genome-wide occupancy profiles of endogenous Myt1l in E13.5 mouse brains (n = 2) or overexpressed Myt1l with (n = 3) or without (n = 2) Ascl1 and Brn2 in MEFs two days after reprogramming. Corresponding regions are displayed across all data sets ± 2 kb from summits. b, Chromatin accessibility based on MNase-seq signal in MEFs25 shows binding enrichment of Myt1l in open and ASCL1 in closed regions. c, Read densities of ASCL1 and BRN2 chromatin binding8 shows minor overlap at Myt1l bound regions. d, The Myt1l AAGTT-core motif (green arrow) is significantly enriched at bound sites across data sets. P-value reported, E-value is 9.6e-3. e, Pie chart indicates enrichment of high confidence Myt1l bound sites at gene promoters.
