Fig. 3: Characterisation of neurogenic and repressive Myt1l domains.
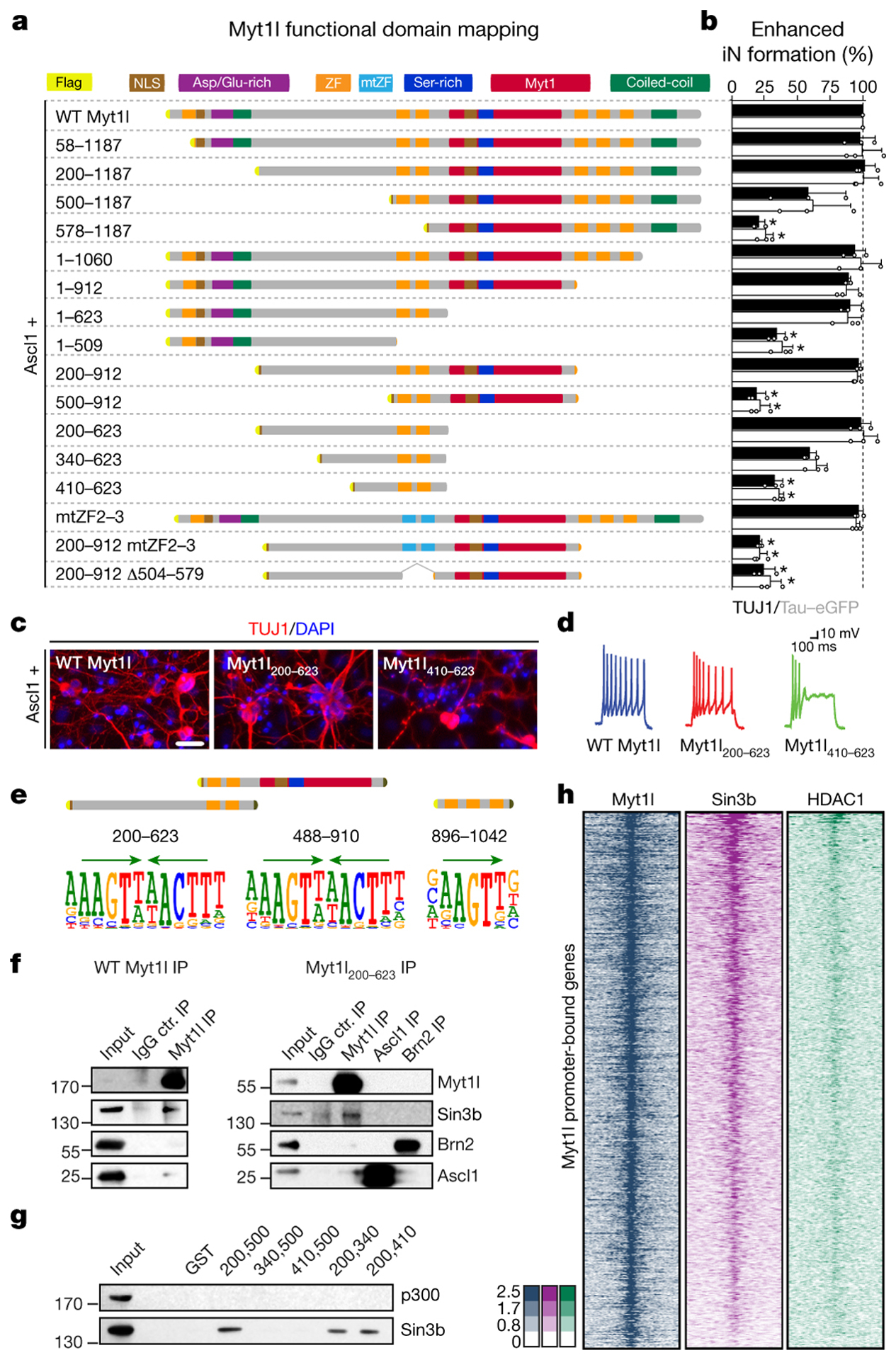
a, Truncation and mutation screen identifies Myt1l domains essential for induced neurogenesis from MEFs upon reprogramming for 14 days with Ascl1. Highlighted are nuclear localisation signals (NLS), aspartic acid/glutamic acid-rich (Asp/Glu-rich), serine-rich (Ser-rich), MYT1, coiled-coil domains, CCHC-type zinc fingers (ZF) and mutants (mtZF). b, Conversion activity compared to Myt1l wt based on number of TUJ1-positive cells with neuronal morphology (black) or TauEGFP expression (grey). n = 3, error bars = SD, t-test * p < 0.005. c, Representative immunofluorescence of iN cells in A; TUJ1 (red), DAPI staining (blue), scale bar 50 μm. d, Representative action potential (AP) traces of iN cells in A upon maturation for 21 days on mouse glia. e, SELEX DNA binding experiments of Myt1l ZF fragments enrich same Myt1l AAGTT-core motif (green arrows). f, Immunoprecipitation show binding of SIN3b to full length and minimal Myt1l in DNase-treated MEF cell lysates two days after transgene overexpression. g, GST pull down from MEF cell lysates identify minimal SIN3b interaction region within functional Myt1l domain. h, Overlapping ChIP-seq chromatin occupancy profiles of overexpressed Myt1l (left, blue), endogenous SIN3b (middle, violet) and HDAC1 (right, green) at Myt1l promoter target sites in MEFs two days after reprogramming induction. n = 2.
