Abstract
The availability of the human genome sequence allowed us to identify a human complement-related, C1r-like protease gene (c1r-LP) located 2 kb centromeric of the C1r gene (c1r). Compared with c1r, c1r-LP carries a large deletion corresponding to exons 4–8 of c1r. The open reading frame of the C1r-LP cDNA predicts a 50 kDa modular protein displaying 52% amino acid residue identity with the corresponding regions of C1r and 75% identity with a previously described murine C1r-LP. The serine protease domain of C1r-LP, despite an overall similarity with the AGY group of complement serine proteases, has certain structural features characteristic of C2 and factor B, thus raising interesting evolutionary questions. Northern blotting demonstrated the expression of C1r-LP mRNA mainly in the liver and ELISA demonstrated the presence of the protein in human serum at a concentration of 5.5±0.9 μg/ml. Immunoprecipitation experiments failed to demonstrate an association of C1r-LP with the C1 complex in serum. Recombinant C1r-LP exhibits esterolytic activity against peptide thioesters with arginine at the P1 position, but its catalytic efficiency (kcat/Km) is lower than that of C1r and C1s. The enzymic activity of C1r-LP is inhibited by di-isopropyl fluorophosphate and also by C1 inhibitor, which forms stable complexes with the protease. Most importantly, C1r-LP also expresses proteolytic activity, cleaving pro-C1s into two fragments of sizes identical with those of the two chains of active C1s. Thus C1r-LP may provide a novel means for the formation of the classical pathway C3/C5 convertase.
Keywords: C1 inhibitor, classical complement pathway, C1r-like protease, human complement-related protein, pro-C1s, serine esterase
Abbreviations: Boc, t-butoxycarbonyl-; C1 INH, complement 1 inhibitor; C1r-LP, C1r-like protease; CCP, complement control protein; CHO, Chinese-hamster ovary; CS, connecting segment; CUB, C1r/C1s, sea urchin protein uEGF, bone morphogenic protein; EGF, epidermal growth factor; SP, serine protease; MASP, mannan-binding lectin-associated SP; iPr2P-F, di-isopropyl fluorophosphate; NHS, normal human serum; ORF, open reading frame; poly(A)+ RNA, polyadenylated RNA; SBzl, thiobenzyl; UTR, untranslated region; Z, benzyloxycarbonyl
INTRODUCTION
The complement system comprises more than 35 plasma and cell surface proteins and represents a major element of innate immunity [1,2]. The system is activated through one of three pathways termed classical, lectin and alternative. The classical pathway is initiated by the binding of the C1 complex to immune complexes or to other activators, such as C-reactive protein complexes and certain pathogens [3,4]. C1 is a macromolecular complex comprising a recognition subunit, C1q and a catalytic Ca2+-dependent tetramer C1s-C1r-C1r-C1s, consisting of two SPs (serine proteases) in their zymogen form [5]. The binding of C1q to an activator leads to conformational changes, which induce the autocatalytic activation of pro-C1r by the cleavage of an Arg–Ile peptide bond [6]. Activated C1r activates pro-C1s, which in turn activates C4 and C2 leading to the formation of the C3 convertase, C4b2a [7,8]. C1r and C1s also express esterolytic activity against synthetic ester substrates containing a P1 arginine or lysine residue [9,10]. Their activity is regulated by the serpin C1 INH (complement 1 inhibitor) [11], which also inactivates some other blood proteases, including factor XIa [12], factor XIIa [13], kallikrein [14] and the MASPs (mannan-binding lectin-associated serine proteases) [15]. It has been reported that C1 INH also prevents the activation of pro-C1r and -C1s by weak activators [16].
C1r is a modular SP that comprises an N-terminal CUB (C1r/C1s, sea urchin protein uEGF, bone morphogenic protein) module [17] followed by an EGF (epidermal growth factor)-like module, a second CUB module, two CCP (complement control protein) modules and a chymotrypsin-like SP domain [18,19]. The CUB1-EGF modules are required for the Ca2+-dependant formation of C1r-C1s heterodimers and the assembly of the C1 complex [20,21]. The CCP1 module is responsible for the dimerization of C1r, whereas the CCP2 module stabilizes the structure of the SP domain and probably enhances its catalytic efficiency [22,23].
Previous work in our laboratory has demonstrated the presence of a novel murine C1r-related gene located approx. 3 kb upstream of the C1rA gene [24]. Compared with the C1rA and C1rB genes, the novel gene displays a large deletion encompassing exons 4–8 and encodes a muC1r-LP (murine C1r-like protein-) that consists of a single CUB module, a truncated CCP-like module, a CS (connecting segment) and an SP domain. muC1r-LP shows 51% overall amino acid identity to corresponding regions of murine C1rA [25]. A search of the GenBank® database for the human orthologue identified a cDNA encoding a human C1r-LP. In this report, we describe the isolation of human C1r-LP cDNA and the production and functional characterization of the recombinant protein.
Part of this work was presented at the XIXth International Complement Workshop, September 22–26, 2002, Palermo, Italy (abstract no. 16 [25a]).
EXPERIMENTAL
C1r-LP cDNA cloning
Total RNA was isolated from human liver as described in [26] and used as template for first strand cDNA synthesis by reverse transcription, using an oligo-dT primer and the Superscript preamplification system (Life Technologies, Gaithersburg, MD, U.S.A.). cDNA encompassing the full-length ORF (open reading frame) of C1r-LP was amplified by PCR using Pfu DNA polymerase (Stratagene, La Jolla, CA, U.S.A.). The forward primer, 5′-ATATCTAGAAAGCTTATGCCTGGACCCAGAGTGTGGG-3′, contained XbaI and HindIII restriction sites and the reverse primer, 5′-TATGCGGCCGCGAATTCTCAATTCTTGCCATTCATCACTCCC-3′, contained NotI and EcoRI restriction sites. All primers were obtained from the Foundation for Research and Technology-Hellas (Heraklion, Greece). The 1496 bp PCR product was digested with HindIII and EcoRI and subcloned into the similarly restricted expression vector pcDNA3 (Invitrogen/Gibco, Carlsbad, CA, U.S.A.). Nucleotide sequencing of the insert on both strands identified it as the C1r-LP ORF described in the GenBank® under accession no. AF178985.
Expression and purification of C1r-LP and production of antiserum
Full-length ORF of C1r-LP cDNA was generated by PCR using the forward primer 5′-TTTGCATGCTCCGTCCTCTTGGCCCAAGAGC-3′ and the reverse primer 5′-TATAAGCTTTCAATTCTTGCCATTCATCACTCCC-3′, containing SphI and HindIII restriction sites respectively. The PCR product was digested with these enzymes and ligated into similarly restricted pQE30 vector (Qiagen, Valencia, CA, U.S.A.), such that the expressed protein would be extended by a His6 tag at its N-terminal end. After verification by sequencing, the construct was expressed in Escherichia coli strain M15 and the recombinant protein was purified from the cleared bacteria lysate by Ni2+-nitrilotriacetate metal-affinity chromatography in denaturing buffers by using the QIA expressionist system (Qiagen). The purified C1r-LP was used to raise antisera in rabbits (Eurogentec, Seraing, Belgium). IgG was isolated from antiserum by caprylic acid precipitation [27] and labelled with horseradish peroxidase (Sigma, Munich, Germany) as described in [28].
For functional assays, recombinant C1r-LP was expressed in CHO-K1 (Chinese-hamster ovary K1) cells (A.T.C.C., Rockville, MD, U.S.A.), by a modification of a previously described procedure [29]. CHO-K1 cells were grown in Ham's F-12 medium supplemented with 10% heat-inactivated FBS and 1% Pen/Strep (Invitrogen/Gibco), at 37 °C in a humidified, 5% CO2 incubator. The cells were transfected with the C1r-LP-pcDNA3 construct by using the TransFast™ transfection reagent (Promega, Madison, WI, U.S.A.), comprising a synthetic cationic lipid and the neutral lipid, dioleoylphosphatidylethanolamine. Construct DNA (15 μg) was used for transfection, in a 1:1 ratio with the transfection reagent, according to the manufacturer's instructions. Selection of neomycin-resistant cells started 72 h after transfection, using 1 mg/ml Geneticin sulphate (G418; Cellgro, Herndon, VA, U.S.A.). After 7 days, G418-resistant cells were subcloned by limiting dilution at 0.8 cell/well in 96-well tissue culture plates. After 10–12 days in culture, supernatants were tested for C1r-LP production by a sandwich ELISA. Anti-C1r-LP IgG was used in the solid phase of the assay, peroxidase-conjugated anti-C1r-LP IgG as the second antibody, and o-phenylenediamine (Sigma) as substrate. Colour development was measured at 490 nm. The highest producer clone was selected, expanded and adapted to large-scale production by growing in 175 cm2 flasks (Sarstedt, Nümbrecht, Germany). Cells were gradually adapted to the serum- and protein-free medium, chemically defined CHO (CD CHO; Invitrogen/Gibco), supplemented with 0.125 mM glutamine. On day 4 of culture, 1 litre of supernatant from the stably transfected cell line, was harvested, centrifuged at 20000 g and concentrated to 50 ml by ultrafiltration. After dialysis against 20 mM Na2HPO4 buffer (pH 8.6) containing 5 mM EDTA, the sample was applied to a MonoQ 10/10 column of an FPLC apparatus (Amersham Biosciences, Freiburg, Germany), equilibrated with the same buffer. A gradient to 0.5 M NaCl in starting buffer was used to elute bound protein.
Western-blot analysis
Protein samples were subjected to SDS/PAGE, under reducing conditions and then transferred on to nitrocellulose membranes. Proteins were detected by rabbit or sheep antisera as indicated followed by the appropriate peroxidase-conjugated anti-IgG (Sigma). Blots were developed by using either chemiluminescent reagents (ECL® Plus kit; Amersham Biosciences) or 4-chloro-1-naphthol (Sigma).
Northern-blot analysis
The multiple tissue Northern blot (ClonTech, Palo Alto, CA, U.S.A.), containing 1 μg of purified poly(A)+ RNA (polyadenylated RNA) from each of twelve human tissues was used to assess expression of C1r-LP in comparison with C1r. cDNA probes were labelled with 32P and used for hybridization according to the manufacturers' instructions. The C1r-LP probe was a 612-bp PCR product encoding the CUB, CCP and CS of the protein. The C1r probe was a 896-bp PCR product encoding the EGF, CUB2, CCP1 and CCP2 modules of C1r; it was amplified from total human liver RNA by using 5′-GATGAATGTGCTTCCCGGAGCAAA-3′ and 5′-CTCTCCCTTCTGTTCATTCTTCCAA-3′ as forward and reverse primers respectively. Images were developed using a phosphor screen, the STORM 860 system and The ImageQuant software for Windows NT (Molecular Dynamics, Sunnyvale, CA, U.S.A.).
ELISA for C1r-LP in serum
The concentration of C1r-LP in human serum was measured by an ELISA, using a rabbit antiserum raised against purified recombinant C1r-LP from CHO-K1 cells. The IgG fraction of the antiserum [27] was used to coat microtitre wells at a concentration of 30 μg/ml. The same IgG anti-C1r-LP labelled with horseradish peroxidase [28] was used for detection at a concentration of 3.3 μg/ml. The assay was developed using o-phenylenediamine as substrate and measuring colour development at 490 nm. Purified recombinant C1r-LP produced by CHO-K1 cells was used to construct the standard curve and the lower limit of detection was 12 ng/ml serum. Human sera from 20 healthy adults were tested in duplicate at a 1/10 dilution.
Immunoprecipitation experiments
To investigate the possible association of C1r-LP with the C1 complex in serum, we precipitated C1q or C1r-LP from NHS (normal human serum) using specific antisera and analysed the resulting precipitates by Western blotting. Briefly, 30 μl aliquots of NHS were mixed with protein G Streptococcus sp. group G cells (Sigma) precoated with rabbit anti-C1r-LP or sheep anti-C1q IgG (The Binding Site, Birmingham, U.K.). The cells were suspended in 100 μl of PBS (pH 7.4) containing 0.5% Triton X-100, 0.25% deoxycholate (Applichem, Darmstadt, Germany) and 0.5 mM CaCl2 or 10 mM EDTA (incubation buffer). After overnight incubation at 4 °C with continuous rotation, the cells were washed three times with 1 ml of corresponding incubation buffer containing 1% SDS (wash buffer) and finally resuspended in 30 μl SDS/PAGE loading buffer and incubated for 3 min at 95 °C. Triplicate 10 μl aliquots of each incubation mixture were subjected to SDS/PAGE followed by Western blotting. Rabbit anti-C1r-LP, sheep anti-C1q or sheep anti-C1s (The Binding Site) and peroxidase-conjugated species-specific anti-IgG antibodies were used to develop the blots. Controls included NHS, NHS incubated with protein G cells or protein G cells coated with preimmune serum and protein G cells incubated with antiserum. To coat with antiserum IgG, 100 μl of protein G cells (10% suspension) were washed three times with wash buffer, resuspended in 100 μl of incubation buffer, mixed with 1 μl of anti-C1q or 6 μl of anti-C1r-LP and incubated for 2 h at 4 °C with continuous rotation. The cells were then washed three times as above and resuspended in 100 μl of incubation buffer.
Esterolytic and proteolytic assays
The esterolytic activity of recombinant C1r-LP was measured using Z-Gly-Arg-SBzl (where Z stands for benzyloxycarbonyl and SBzl for thiobenzyl) or Boc-Gly-Leu-Ala-Arg-SBzl (where Boc stands for t-butoxycarbonyl; Richman Chemical, Lower Gwynedd, PA, U.S.A.) as substrate. Assays were performed in microplate wells by a modification of the method described by Kam et al. [30]. C1r-LP (128–320 nM) purified from CHO cell supernatants was added to 0.08–0.8 mM thioester and 1.6 mM Ellman's reagent [5,5-dithiobis-(2-nitrobenzonic acid); Sigma] in 250 μl of 0.1 M Hepes (pH 7.5) containing 0.5 M NaCl and 16% Me2SO. The release rate of HS-Bzl was measured kinetically for 15 min at 25 °C as absorbance at 405 nm on a Vmax kinetic microplate reader (Molecular Devices, Menlo Park, CA, U.S.A.). Activated C1r (23 nM) and C1s (14 nM) were obtained from Calbiochem–Novabiochem (San Diego, CA, U.S.A.) and recombinant Bb fragment of factor B (0.11–0.38 μM) was produced and purified as described in [31] and assayed under the same conditions as positive controls. Kinetic constants were calculated by the Lineweaver–Burk method based on at least five substrate concentrations. Correlation coefficients in all cases were >0.99. The same assay was performed using mixtures of C1r-LP and C1 INH (Sigma) preincubated at 25 °C for 45 min at molar ratios of 1:1 or 1:3 or with C1r-LP preincubated at 25 °C for 45 min with 5 or 0.5 mM iPr2P-F (di-isopropyl fluorophosphate; Calbiochem–Novabiochem).
For proteolytic assays, the indicated amounts of purified recombinant C1r-LP and substrate protein were incubated at 30 °C for 30 min in PBS (pH 7.2) and the products were analysed by SDS/PAGE on 10% acrylamide gels. C1s proenzyme was purchased from The Binding Site and C3 from Calbiochem–Novabiochem.
RESULTS
C1r-LP gene structure
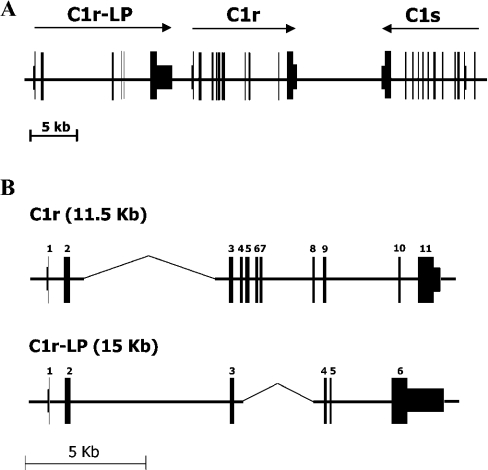
A map of the 50 kb region on chromosome 12p13 encompassing the genes encoding C1r-LP (c1r-lp), C1r (c1r) and C1s (c1s) was constructed on the basis of data from GenBank® files (accession nos. NT_009714, NT_009759, AB083037) (Figure 1A). As shown, c1r-lp and c1r are at a distance of approx. 2 kb from each other and in the same transcriptional orientation, whereas c1s lies approx. 9 kb telomeric of c1r and in opposite transcriptional orientation. c1r-lp consists of six exons spanning 14.8 kb of DNA. The first exon encodes the 5′-UTR (untranslated region) and part of the leader peptide, whereas exon 6 encodes a SP domain uninterrupted by introns and a large approx. 1.9 kb 3′-UTR. c1r spans 11.5 kb of DNA and comprises 11 exons (Figure 1B) [32]. Comparison of the two genes indicates that they exhibit similar overall organization, except that the region corresponding to exons 4–8 of c1r is missing from c1r-lp, perhaps having been deleted. Despite the deletion, c1r-lp is larger than c1r, due to the large size of its intron 2.
Figure 1. The human C1r-LP/C1r/C1s gene cluster.
(A) Map of the approx. 50 kb region on chromosome 12p13 comprising the c1r-LP, c1r and c1s genes. Arrows indicate the direction of transcription. (B) Comparison of c1r-LP and c1r. The overall organization of the two genes is similar, but the region corresponding to exons 4–8 of c1r has been deleted in c1r-LP. Gaps (indicated by thinner lines) have been introduced in the two genes to facilitate their alignment.
C1r-LP cDNA and predicted amino acid sequence
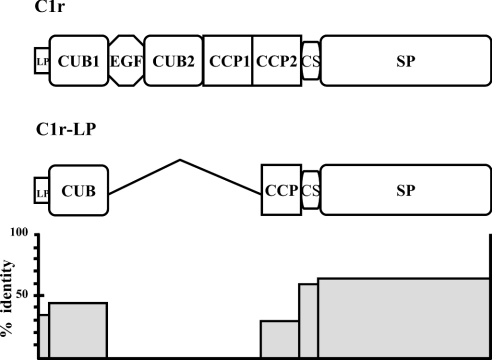
The nucleotide sequence of our 1464 bp C1r-LP cDNA clone consists of the entire ORF, including 120 bp at the 5′-end encoding a putative 40 amino acid long leader peptide. A second ATG codon is present 69 nt downstream of the first one and could represent the main translational initiation site. The sequence is identical with the previously reported C1r-LP cDNA clone (accession no. AF178985), except for four silent nucleotide substitutions, A285G, C414T, T456C and T468C, which could represent polymorphisms. The C1r-LP nucleotide sequence and the predicted amino acid sequence display 55 and 52% identity respectively to the corresponding regions of C1r and 75 and 75.2% identity respectively to muC1r-LP. Finally, human C1r-LP displays 26% amino acid residue identity to C1s. The predicted polypeptide chain of mature C1r-LP has a calculated molecular mass of 50 kDa and displays seven potential N-linked glycosylation sites. Comparison with C1r (Figure 2) indicates that the amino acid sequence of C1r-LP consists of a 40 or 17 residues long leader peptide, followed by a 123 residues long CUB module, a CCP-like module of 67 amino acids, a CS of 14 amino acids and a SP domain of 243 amino acids.
Figure 2. Comparison of the modular structures of C1r-LP and C1r.
Compared with C1r, C1r-LP lacks the EGF, the second CUB and the first CCP modules. In addition, the CCP module of C1r-LP is truncated, missing the first cysteine of CCPs. The histogram at the bottom shows the percentage of amino acid residue identity between modules of C1r-LP and C1r. LP, leader peptide.
The CUB domain of C1r-LP has conserved the ‘signature’ residues of the domain, including the two highly conserved cysteine residues. It appears to belong to the Ca2+-binding subgroup of CUB domains, which includes C1r, C1s and the MASPs [33], as it displays three of the four conserved residues that co-ordinate the ion, whereas a glutamine substitutes the second conserved aspartic acid. In contrast, the CCP-like module, particularly its N-terminal region, is the least identical segment of C1r-LP, with C1r displaying 26% residue identity with CCP2 of C1r, whereas the leader peptide, CUB, CS and SP domain display 35, 49, 56 and 62% identity with the corresponding modules of C1r (Figure 2). Most importantly, the CCP-like module is missing the first of the four highly conserved cysteine residues, which in typical CCP modules is disulphide-bonded to the third cysteine residue [34].
The SP domain of C1r-LP has several interesting structural characteristics. Similar to C1r, C1s, MASP2 and MASP3, it is encoded by a single exon and lacks the highly conserved histidine disulphide loop of typical SPs (Figure 3). In addition, similar to other members of the AGY group of SPs, its active site Ser195 (chymotrypsinogen numbering is used for SP domain residues to allow comparisons) is encoded by AGT instead of the typical TCN and a valine instead of the conserved proline is present at position +3 relative to Ser195. The highly conserved catalytic triad residues Asp102-His57-Ser195 and the three residues, Ser-Trp-Gly214–216 forming the non-specific substrate-binding site of SPs are present in C1r-LP. However, C1r-LP is lacking the typical arginine or lysine residue at the cleavage/activation site of SPs and the conserved N-terminal amino acid sequence that plays a critical role in the activation of SPs. This feature is not unique to C1r-LP but also characterizes factor B and C2, the SP domain of which remains linked to their preceding von Willebrand factor type A domain. In addition, like factor B and C2, C1r-LP has a non-charged residue at position 189 corresponding to the bottom of the primary substrate specificity pocket (S1 site) instead of the aspartic acid of C1r and all other SPs with trypsin-like specificity. C1r-LP and C2 have serine residues at that position, whereas factor B has an asparagine residue [35]. Finally, like factor B and C2, C1r-LP has an aspartic residue at position 226, instead of the highly conserved glycine of typical SPs.
Figure 3. Amino acid sequence alignment of SP domains of chymotrypsin, trypsin, C1r, C1r-LP and factor B.
Numbering at the top is for chymotrypsinogen. Arrows indicate the catalytic triad residues, stars indicate residue 189 at the bottom of the S1 pocket and residue 226, which is aspartic residue in C1r-LP and factor B. The conserved newly formed N-terminal sequence of active SPs is indicated by shading. CHM, bovine chymotrypsin; TRP, bovine trypsin; fB, complement factor B.
C1r-LP is a serum protein of hepatic origin
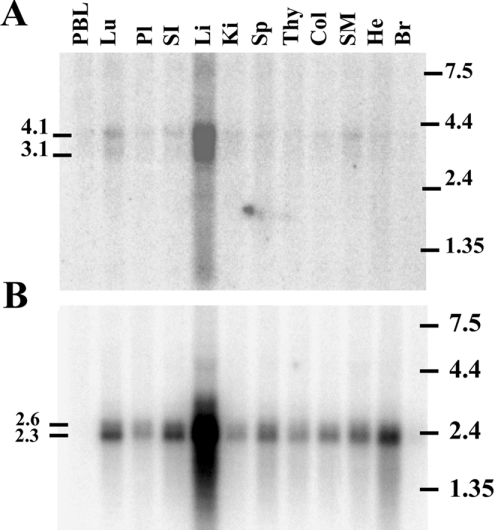
Northern-blot analysis of poly(A)+ RNA from several human tissues using a C1r-LP-specific probe revealed the presence of two bands of approx. 3.1 and 4.1 kb mainly in the liver. Fainter signals of these bands were also detected in the lung, kidney, small intestine and skeletal muscles (Figure 4A).
Figure 4. Tissue distribution of C1r-LP (A) and C1r (B) mRNAs.
Northern-blot analysis of poly(A)+ RNA from 12 human tissues. The blot was probed with a 612 bp C1r-LP cDNA probe encoding the CUB, CCP and CS region of the protein and with a 896 bp C1r probe encoding the EGF, CUB2, CCP1 and CCP2 region of the protein. PBL, peripheral blood leukocytes; Lu, lung; Pl, placenta; SI, small intestine; Li, liver; Ki, kidney; Sp, spleen; Thy, thymus; Col, colon; SM, skeletal muscle; He, heart; Br, brain.
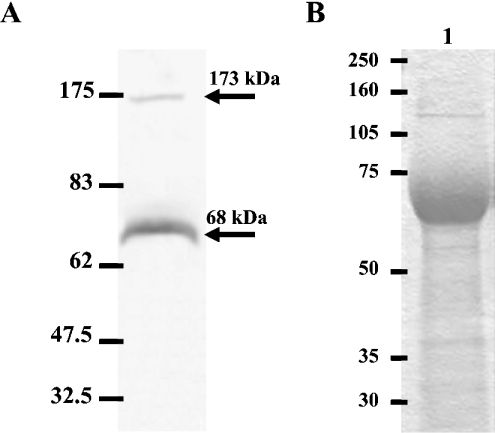
The C1r-specific probe also detected a double band of approx. 2.6 and 2.3 kb mainly in the liver, but also in several other tissues, including lung, small intestine, spleen, colon, skeletal muscles and heart (Figure 4B). The large size of the C1r-LP mRNA is probably due to its very long 3′-UTR, whereas the presence of multiple polyadenylation signals could possibly explain the double band. To investigate the possible secretion of C1r-LP from the liver into the blood, we performed Western-blot analysis of NHS using the anti-C1r-LP serum. As shown (Figure 5A), C1r-LP was detected as a band of 68 kDa. The difference from the calculated 50 kDa of the polypeptide chain is probably due to glycosylation of all or most of the seven potential N-glycosylation sites. This interpretation is supported by the similar size of the recombinant protein expressed in CHO cells (Figure 5B). An additional faint band of 175 kDa was also present in the Western blot. The identity of this band is currently unknown, but based on its size it could perhaps represent the complex of C1r-LP with C1 INH (see below).
Figure 5. Western blotting and SDS/PAGE of C1r-LP.
(A) Western-blot analysis of NHS using a rabbit anti-C1r-LP. The blot was developed using a horseradish peroxidase-conjugated anti-rabbit IgG, followed by 4-chloro-1-naphthol. The calculated molecular mass of detected bands is shown on the left and the position of molecular mass markers on the right. (B) SDS/PAGE (10% gel) under reducing conditions of purified recombinant C1r-LP expressed in CHO-K1 cells. Positions of molecular mass markers are on the left.
The mean±S.D. concentration of C1r-LP in NHS was 5.5±0.9 μg/ml, as determined by ELISA using 20 sera from healthy human adults.
C1r-LP does not associate with the C1 complex
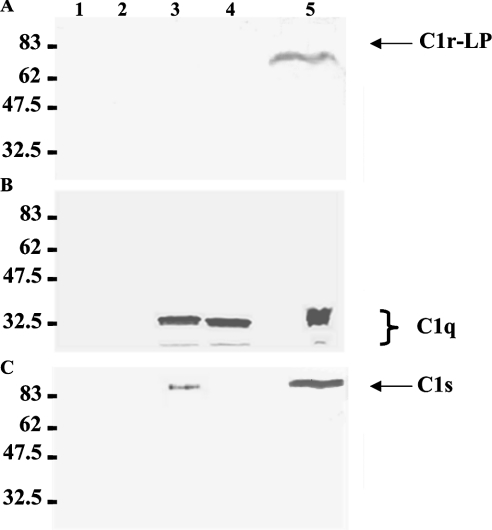
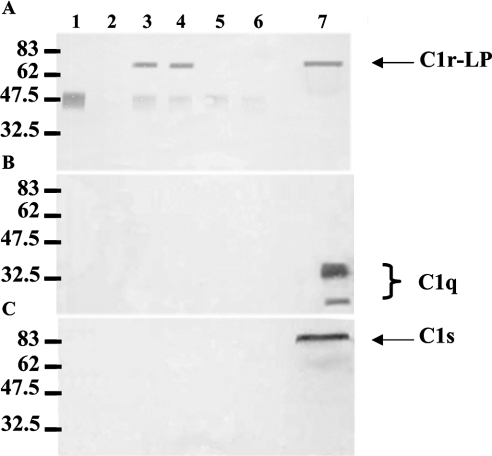
To examine the possible association of C1r-LP with the C1 complex in serum, we precipitated C1q or C1r-LP from NHS using specific antisera and analysed the immunoprecipitates by Western blotting for the presence of C1r-LP, C1q or C1s. Figure 6 shows three Western blots of anti-C1q immunoprecipitates. C1q is detected (Figure 6B) as a triple band in precipitates obtained in the presence of either Ca2+ (lane 3) or EDTA (lane 4). C1q is also detected in the NHS control (lane 5), but not in the negative control lacking NHS (lane 1) or anti-C1q serum (lane 2). As expected, C1s co-precipitated with C1q from NHS in the presence of Ca2+ (Figure 6C, lane 3), but not in the presence of EDTA (lane 4). C1r-LP did not co-precipitate with C1q under either of the conditions (Figure 6A). The lack of association of C1r-LP with the C1 complex was also indicated by the converse experiment shown in Figure 7. Neither C1q (Figure 7B) nor C1s (Figure 7C) was detected in immunoprecipitates of C1r-LP from NHS (Figure 7A). The approx. 50 kDa band seen in the anti-C1r-LP blot is apparently due to the heavy chain of the rabbit anti-C1r-LP IgG, since it is also present in the mixture lacking NHS (lane 1) and when preimmune rabbit serum was used instead of anti-C1r-LP serum (lanes 5 and 6).
Figure 6. Western-blot analysis of C1q immunoprecipitates from NHS.
C1q was precipitated from NHS using a rabbit antiserum and protein G cells. Triplicate aliquots of the precipitate were subjected to SDS/PAGE (10% gel) under reducing conditions, followed by Western blotting using antisera against C1r-LP (A), C1q (B) or C1s (C). Protein bands were developed using species-specific horseradish peroxidase-conjugated anti-IgG sera and 4-chloro-1-naphthol. Lane 1, anti-C1q+protein G control; lane 2, NHS+protein G control; lane 3, NHS+anti-C1q+protein G, 0.5 mM Ca2+; lane 4, NHS+anti-C1q+protein G, 10 mM EDTA; lane 5, NHS.
Figure 7. Western-blot analysis of C1r-LP immunoprecipitates from NHS.
C1r-LP was precipitated from NHS using a rabbit antiserum and protein G cells. Triplicate aliquots of the precipitate were subjected to SDS/PAGE (10% gel) under reducing conditions, followed by Western blotting using antisera against C1r-LP (A), C1q (B) or C1s (C). Protein bands were developed using species-specific horseradish peroxidase-conjugated anti-IgG sera and 4-chloro-1-naphthol. Lane 1, anti-C1r-LP+protein G control; lane 2, NHS+protein G control; lane 3, NHS+anti-C1r-LP+protein G, 0.5 mM Ca2+; lane 4, NHS+anti-C1r-LP+protein G, 10 mM EDTA; lane 5, NHS+preimmune rabbit serum+protein G, 0.5 mM Ca2+; lane 6, NHS+preimmune rabbit serum+protein G, 10 mM EDTA; lane 7, NHS.
Esterolytic activity of recombinant C1r-LP
C1r-LP expressed in CHO-K1 cells was purified from culture supernatants by FPLC on monoQ columns. On SDS/PAGE (10% gel) purified recombinant C1r-LP migrated as a 68 kDa protein (Figure 5B). The esterolytic activity of recombinant C1r-LP was assayed using Z-Gly-Arg-SBzl and Boc-Gly-Leu-Ala-Arg-SBzl as substrates. These thioesters have been shown to be good substrates for C1r and C1s [10]. Boc-Gly-Leu-Ala-Arg-SBzl, which mimics the cleavage/activation site of C3, is also hydrolysed by the Bb fragment of factor B [30]. For comparison, C1r and C1s were assayed against the first and Bb against the second thioester under the same conditions as C1r-LP. Kinetic constants for the hydrolysis of the two substrates by the four enzymes are summarized in Table 1. As shown, C1r-LP displays enzymic activity against both substrates. Its catalytic efficiency (kcat/Km) against Z-Gly-Arg-SBzl is low, approx. 5- and 30-fold lower than that of C1r and C1s respectively although 2-fold higher than Bb against Boc-Gly-Leu-Ala-Arg-SBzl. An additional experiment (results not shown) demonstrated that C1r-LP also hydrolysed Ac-Gly-Lys-OMe (where Ac stands for acetyl), which is also a substrate for C1r. Hydrolysis of the methyl ester by C1r-LP was again less efficient than C1r.
Table 1. Kinetic constants for hydrolysis of the synthetic thioester substrates by C1r-LP, C1r, C1s and fragment Bb.
Esterolytic activity was measured by adding C1r-LP (128–320 nM), C1r (23 nM), C1s (14 nM) or Bb (110–380 nM) to 0.08–0.8 mM thioester and 1.6 mM Ellman's reagent in 250 μl of 0.1 M Hepes (pH 7.5), 0.5 M NaCl, 16% Me2SO, at 25 °C. Release rate of HS-Bzl was measured kinetically at 405 nm. Kinetic constants were calculated by the Lineweaver–Burk method.
| Z-Gly-Arg-SBzl | Boc-Gly-Leu-Ala-Arg-SBzl | |||||
|---|---|---|---|---|---|---|
| Enzyme | kcat/Km (s−1·M−1) | kcat (s−1) | Km (mM) | kcat/Km (s−1·M−1) | kcat (s−1) | Km (mM) |
| C1r-LP* | 1×103±0.1×103 | 1.8±0.1 | 2.0±0.3 | 0.2×103±0.02×103 | 0.1±0.01 | 0.6±0.1 |
| C1r* | 5×103±0.3×103 | 2.0±0.5 | 0.4±0.01 | − | − | − |
| C1s† | 26×103 | 55.1 | 2.1 | − | − | − |
| rBb† | − | − | − | 0.1×103 | 0.7 | 5.6 |
*The values of individual parameters are means±S.E.M. for four independent determinations.
†The values of individual parameters are the average of two independent determinations.
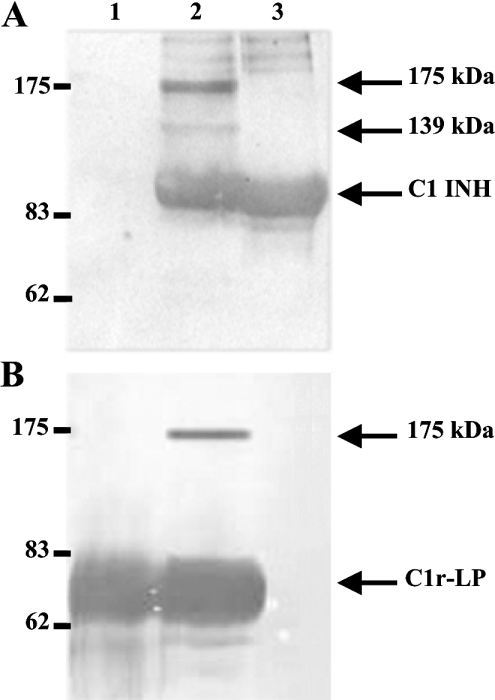
Hydrolysis of Z-Gly-Arg-SBzl by C1rLP was inhibitable by C1 INH. When C1r-LP was incubated with C1 INH at 1:1 or 1:3 molar ratios, its catalytic efficiency (kcat/Km) became approx. 5-fold lower or was completely inhibited respectively. To investigate whether inhibition was associated with the formation of stable C1r-LP–C1 INH complexes, the two proteins were incubated at a 1:1 molar ratio for 1 h at 37 °C. Complex formation was assessed by SDS/PAGE (7.5% gel), under reducing conditions, followed by Western blotting. As shown in Figure 8, both anti-C1r-LP and anti-C1 INH sera detected in addition to their respective uncomplexed proteins, a band of 175 kDa, which apparently represents a stable complex of the two proteins. In addition, a band of 139 kDa is detected only by the anti-C1 INH serum. The identity of this band is currently unknown. Finally, the esterolytic activity of C1r-LP measured against Boc-Gly-Leu-Ala-Arg-SBzl was completely inhibited by 5 mM iPr2P-F, whereas 0.5 mM iPr2P-F produced approx. 70% inhibition of activity.
Figure 8. Western blots of C1r-LP/C1 INH mixtures.
Purified recombinant C1r-LP was incubated with an equimolar amount of C1 INH at 37 °C for 1 h and duplicate aliquots of the mixture were subjected to SDS/PAGE (7.5% gel) under reducing conditions, followed by Western blotting. The upper blot (A) was developed with anti-C1 INH and the lower blot (B) with anti-C1r-LP serum. Both blots were developed with peroxidase-conjugated anti-rabbit IgG, followed by chemiluminescent reagents (A) or 4-chloro-1-naphthol (B). Lane 1, purified C1r-LP; lane 2, C1r-LP/C1 INH mixture; lane 3, C1 INH. Positions of molecular mass markers are on the left. Arrows indicate the uncomplexed proteins and the two observed complexes of 175 and 139 kDa.
C1r-LP specifically cleaves pro-C1s
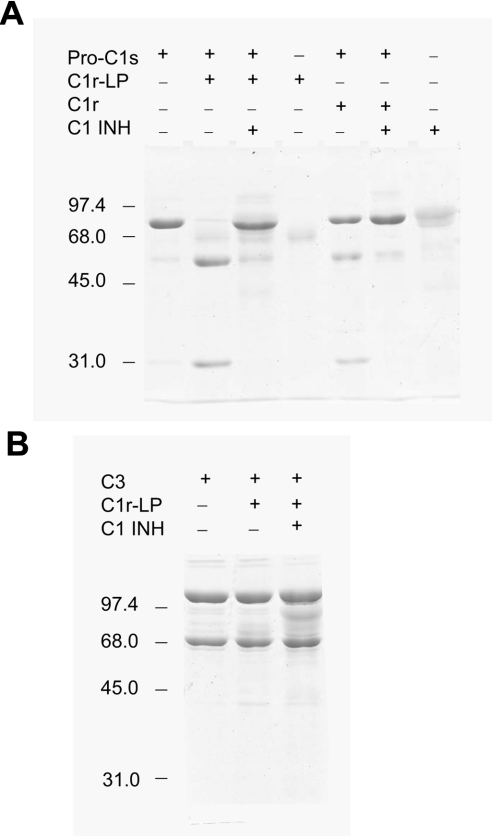
The proteolytic activity of C1r-LP was tested against pro-C1s, the only known substrate of C1r. As shown in Figure 9, the incubation of pro-C1s with C1r-LP at a 1:2 enzyme to substrate molar ratio converted the single 80 kDa polypeptide chain of pro-C1s into a two-chain structure. The sizes of the two chains, estimated at approx. 52 and 28 kDa, are identical with those of the A and B chain respectively of C1s produced by cleavage of pro-C1s by C1r. This result suggests that similar to C1r, C1r-LP can convert pro-C1s into active C1s. C1 INH at a 4-fold molar excess over C1r-LP inhibited almost completely the cleavage of pro-C1s. Incubation of C3, used as control, with the same concentration of C1r-LP under the same conditions, resulted in minimal breakdown of C3 into two fragments of approximate sizes of 45 and 42 kDa. However, C1 INH did not inhibit cleavage of C3, suggesting that it was not catalysed by C1r-LP, but by a trace of contaminating protease perhaps derived from the CHO cells. Factor B also used as control was not cleaved by C1r-LP (results not shown).
Figure 9. Proteolytic activity of C1r-LP.
(A) Purified recombinant C1r-LP (1.75 μg) was incubated at 30 °C for 30 min with pro-C1s (4 μg) in 15 μl of PBS (pH 7.2) in the absence or presence of C1 INH (7.8 μg) and the products were analysed by SDS/PAGE (10% gel). (B) C3 (4 μg) was incubated as in (A) with C1-LP (1.75 μg) in the absence or presence of C1 INH (7.8 μg) and the products were analysed by SDS/PAGE (10% gel).
DISCUSSION
In the present study, we describe a novel human complement-related protease, C1r-LP, homologous to complement component C1r. C1r-LP is encoded by a gene containing a large deletion compared with c1r and is located in close proximity to c1r on chromosome 12p13. As a result, the encoded protein is truncated in comparison with C1r, comprising single CUB and CCP-like modules followed by a CS and an SP domain. c1r-lp is expressed mainly in the liver and the protein is secreted in the blood. C1r-LP is heavily glycosylated and despite certain unique characteristics of its SP domain, expresses trypsin-like serine esterase activity, inhibitable by iPr2P-F and C1 INH. More importantly, C1r-LP also expresses proteolytic activity converting pro-C1s into a two-chain structure, similar to active C1s.
The CUB domain of C1r-LP apparently belongs to the recently described subset of proteins, which contain Ca2+-binding CUB domains [33]. The subset includes C1s, C1r and the MASPs, but also non-complement proteins and is characterized by four conserved residues (Tyr17, Glu45, Asp53 and Asp98 in C1s). The crystal structure of the N-terminal fragment of C1s, comprising the CUB1 and EGF-like domains, has demonstrated that the three acidic residues are directly involved in the co-ordination of the Ca2+, whereas Tyr17 is closely associated with the ion-binding site. The bound Ca2+ stabilizes the CUB structure and may play a role in the assembly of the macromolecular C1qC1r2C1s2 complex [33]. C1r-LP has retained at the proper positions three of the four conserved amino acids, whereas a glutamine is substituted for the second aspartic acid. It thus appears probable that C1r-LP can also bind Ca2+. These considerations raised the possibility that C1r-LP may be associated with the C1 complex in serum, perhaps playing a regulatory role. However, immunoprecipitation experiments (Figures 6 and 7) failed to demonstrate such an association.
The SP domain of C1r-LP shares structural features with two groups of complement enzymes, C1r/C1s/MASP-2 and -3 on the one hand and C2/factor B on the other. Similar to the enzymes of the first group, the SP domain of C1r-LP is encoded by an intronless exon, is missing the highly conserved ‘histidine disulphide loop’ and its catalytic Ser195 is encoded by AGY. Although these similarities would be expected since c1r-LP and c1r probably are gene-duplication products, the structural similarities with factors B and C2 are surprising, because the two groups of complement SPs are believed to have evolved separately for a very long evolutionary time with their unique structural characteristics being established in the jawed vertebrate lineage before the divergence of the chondrichthyes [36]. Yet, like C2, C1r-LP has a serine at the position corresponding to residue 189 of chymotrypsin at the bottom of the S1 pocket, whereas all SPs with trypsin-like substrate specificity have an aspartic residue at that position. Factor B also has a non-charged residue, asparagine, at that position. In addition, like C2 and factor B, C1r-LP has an aspartic residue at position 226 of chymotrypsin, instead of the highly conserved glycine of typical SPs. In trypsin-like SPs, Asp189 plays a fundamental role in positioning the P1-Arg or -Lys of the substrate for nucleophilic attack by Ser195. Mutational and structural [31,35] analyses have demonstrated that the loss of the negative charge in the S1 pocket of factor B is compensated by the presence of Asp226, which is the primary structural determinant for P1-Arg binding. The presence of an aspartic residue in the corresponding position of C1r-LP is unique among the members of the C1r/C1s/MASPs group and suggests a structural arrangement of the S1 pocket similar to that of factor B.
From a functional point of view, the absence of a cleavage/activation arginine or lysine peptide bond probably is more significant. In typical SPs transition from the zymogen to the active protease form is triggered by the cleavage of an arginine or more rarely lysine bond, which results in the generation of a new positively charged and highly conserved N-terminal sequence. The latter is inserted into the hydrophobic region adjacent to the S1 pocket and forms an ion bond with the highly conserved Asp194, forcing the catalytic centre to adopt its active conformation [37]. In C1r-LP, a threonine is substituted for the arginine at the cleavage/activation site of C1r (Figure 3) and the conserved Ile-Ile-Gly-Gly- sequence that forms the newly generated N-terminal of C1r has been replaced by Thr-Leu-Gly-Ser-. The SP domains of factor B and C2 also lack a free N-terminus, being connected to a von Willebrand factor type A domain. In the absence of a cleavage/activation site, an alternative activation mechanism must exist. In factor B, the active conformation of the catalytic centre is supposed to be induced by the cofactor C3b and/or by the substrate C3 [31]. The absence of a typical cleavage/activation site and the low catalytic efficiency of C1r-LP against peptide thioesters (Table 1) suggest that its active centre is in inactive conformation. This implies that a presently unknown mechanism induces the active conformation.
The reactivity of C1r-LP with inhibitors is consistent with its low esterolytic activity. Complete inhibition required a high, 5 mM, concentration of iPr2P-F and a 3-fold molar excess of C1 INH. However, the mechanism of inhibition by the serpin appears to be similar to that of other SPs, including the formation of a stable, apparently covalent complex (Figure 8). The formation of this complex is consistent with the expression of serine esterase activity by C1r-LP, although only a small percentage of C1r-LP was engaged in covalent complex formation at an enzyme-to-inhibitor ratio causing 5-fold decrease in catalytic efficiency. The discrepancy could perhaps be explained by hypothesizing that in serum there is an equilibrium between an inactive and an active conformation of C1r-LP, with the former encompassing the majority of the enzyme. C1 INH has two effects: it forms covalent complexes with the active form and non-covalent complexes with the inactive form, preventing it from converting into the active conformation. The latter interaction would be similar to the proposed interaction of C1 INH with pro-C1r [16]. Alternatively, it could be proposed that the inactive form also expresses low-level activity, which is inhibited by the serpin without covalent binding.
The identity of the additional, 139 kDa, C1 INH-containing band cannot be ascertained on the basis of available data. The possibility exists that it represents a complex between C1 INH and a trace SP contaminating the C1r-LP preparation. We consider this possibility unlikely, because C1 INH only reacts with a limited number of enzymes and the recombinant protein was purified from serum-free culture media. C1 INH–SP complexes of size smaller than the sum of the two intact proteins have been reported before and were attributed to cleavage of either C1 INH acting as suicide substrate [38] or of the enzyme, which becomes extremely sensitive to proteolysis when complexed with the serpin [39]. Furthermore, it has been reported that C1r, C1s [40] and kallikrein [41] in complex with C1 INH lose antigenic determinants, becoming completely or partially unreactive with specific antisera. It seems possible that the 139 kDa band represents a complex between a fragment of C1r-LP and C1 INH non-reactive with our anti-C1r-LP serum.
The unique structural features of the SP domain of C1r-LP, particularly the absence of a typical cleavage/activation site combined with its low esterolytic activity and low reactivity with inhibitors made doubtful its potential for expressing proteolytic activity. Nevertheless, as shown in Figure 9 C1r-LP expresses proteolytic activity against pro-C1s, cleaving it into two fragments of size identical with that of the A and B chain of active C1s. It thus appears probable that C1r-LP can activate pro-C1s perhaps leading to the formation of the classical pathway C3/C5 convertase in serum, subject to regulation by C1 INH. In terms of the mechanism of activation, it seems possible that like factor D [42] C1r-LP belongs to the group of SPs that are activated by reversible conformational changes induced by the substrate.
In conclusion, in the present study we have described a novel human SP in blood, C1r-LP, which although missing several interacting modules, is similar to complement protease C1r, particularly for the structure of the SP domain. The full spectrum of its natural substrates and its physiological function remain to be determined; however, C1r-LP expresses catalytic activity against pro-C1s and therefore it could perhaps provide a novel means for activating the classical complement pathway.
While this manuscript was in the final stage of revision, we became aware of a paper by Wicher and Fries [43] published recently. The authors demonstrate intracellular cleavage of prohaptoglobin into haptoglobin by C1r-LP, using co-transfection of cells with the respective cDNAs. They also report that using the same co-transfection system, they failed to observe cleavage of pro-C1s.
Acknowledgments
We thank Dr G. Arlaud, Dr N. Thielens and Dr V. Rossi for help with the esterolytic assays and Dr G. Panayotou for help with protein purification. We also thank the anonymous reviewer who suggested the proteolytic experiments. This work was supported in part by a grant from the General Secretariat of Research and Technology of Greece and by National Institutes of Health grant AR44505.
References
- 1.Volanakis J. E. Overview of the complement system. In: Volanakis J. E., Frank M. M., editors. The Human Complement System in Health and Disease. New York: Marcel Dekker; 1998. pp. 9–32. [Google Scholar]
- 2.Walport M. J. Complement. First of two parts. N. Engl. J. Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 3.Volanakis J. E. Complement activation by C-reactive protein complexes. Ann. N. Y. Acad. Sci. 1982;389:235–250. doi: 10.1111/j.1749-6632.1982.tb22140.x. [DOI] [PubMed] [Google Scholar]
- 4.Loos M. Antibody-indepentent activation of C1, the first component of complement. Ann. Immunol. (Paris) 1982;133:165–179. doi: 10.1016/0769-2625(82)90030-7. [DOI] [PubMed] [Google Scholar]
- 5.Arlaud G. J., Colomb M. G., Gagnon J. A functional model of the human C1 complex. Immunol. Today. 1987;8:106–111. doi: 10.1016/0167-5699(87)90860-7. [DOI] [PubMed] [Google Scholar]
- 6.Arlaud G. J., Gagnon J. Identification of the peptide bond cleaved during activation of human C1r. FEBS Lett. 1985;180:234–238. doi: 10.1016/0014-5793(85)81077-2. [DOI] [PubMed] [Google Scholar]
- 7.Schumaker V. N., Zavodszky P., Poon P. H. Activation of the first component of the complement system. Annu. Rev. Immunol. 1987;5:21–42. doi: 10.1146/annurev.iy.05.040187.000321. [DOI] [PubMed] [Google Scholar]
- 8.Spycher S. E., Nick H., Rickii E. E. Human complement component C1s. Partial sequence determination of the heavy chain and identification of the peptide bond cleaved during activation. Eur. J. Biochem. 1986;156:49–57. doi: 10.1111/j.1432-1033.1986.tb09546.x. [DOI] [PubMed] [Google Scholar]
- 9.Andrews J. M., Baillie R. D. The enzymatic nature of the human C1r: a subcomponent of the first component of complement. J. Immunol. 1979;123:1403–1408. [PubMed] [Google Scholar]
- 10.McRae B. J., Lin T. Y., Powers J. C. Mapping the substrate binding site of human C1r and C1s with peptide thioesters. Development of new sensitive substrates. J. Biol. Chem. 1981;256:12362–12366. [PubMed] [Google Scholar]
- 11.Davis A. E., III C1 inhibitor and hereditary angioneurotic edema. Annu. Rev. Immunol. 1988;6:595–628. doi: 10.1146/annurev.iy.06.040188.003115. [DOI] [PubMed] [Google Scholar]
- 12.Wuillemin W. A., Minnema M., Meijers J. C., Roem D., Eerenberg A. J., Nuijens J. H., Ten Cate H., Hack C. E. Inactivation of factor XIa in human plasma assessed by measuring factor XIa-protease inhibitor complexes: major role for C1-inhibitor. Blood. 1995;85:1517–1526. [PubMed] [Google Scholar]
- 13.Pixley R. A., Schapira M., Colman R. W. The regulation of human factor XIIa by plasma proteinase inhibitors. J. Biol. Chem. 1985;260:1723–1729. [PubMed] [Google Scholar]
- 14.Schapira M., Scott C. F., Colman R. W. Contribution of plasma protease inhibitors to the inactivation of kallikrein in plasma. J. Clin. Invest. 1982;69:462–468. doi: 10.1172/JCI110470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushita M., Thiel S., Jensenius J. C., Terai I., Fujita T. Proteolytic activities of two types of mannose-binding lectin-associated serine protease. J. Immunol. 2000;165:2637–2642. doi: 10.4049/jimmunol.165.5.2637. [DOI] [PubMed] [Google Scholar]
- 16.Ziccardi R. J. A new role for C-1-inhibitor in homeostasis: control of activation of the first component of human complement. J. Immunol. 1982;128:2505–2508. [PubMed] [Google Scholar]
- 17.Bork P., Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J. Mol. Biol. 1993;23:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 18.Journet A., Tosi M. Cloning and sequencing of full-length cDNA encoding the precursor of human complement component C1r. Biochem. J. 1986;240:783–787. doi: 10.1042/bj2400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arlaud G. J., Willis A. C., Gagnon J. Complete amino acid sequence of the A chain of human complement-classical-pathway enzyme C1r. Biochem. J. 1987;241:711–720. doi: 10.1042/bj2410711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai S. W., Poon P. H., Schumaker V. N. Expression and characterization of a 159 aminoacid, N-terminal fragment of human complement component C1s. Mol. Immunol. 1997;34:1273–1280. doi: 10.1016/s0161-5890(97)00149-1. [DOI] [PubMed] [Google Scholar]
- 21.Thielens N. M., Enrié K., Lacroix M., Jaquinod M., Hernandez J. F., Esser A. F., Arlaud G. J. The N-terminal CUB-EGF module pair of human protease C1r binds Ca2+ with high affinity and mediates Ca2+-dependent interaction with C1s. J. Biol. Chem. 1999;274:9149–9159. doi: 10.1074/jbc.274.14.9149. [DOI] [PubMed] [Google Scholar]
- 22.Kardos J., Gal P., Szilagyi L., Thielens N. M., Szilagyi K., Lorincz Z., Kulcsar P., Graf L., Arlaud G. J., Zavodszky P. The role of the individual domains in the structure and function of the catalytic region of a modular serine protease, C1r. J. Immunol. 2001;167:5202–5208. doi: 10.4049/jimmunol.167.9.5202. [DOI] [PubMed] [Google Scholar]
- 23.Budayova-Spano M., Lacroix M., Thielens N. M., Arlaud G. J., Fontecilla-Camps J. C., Gaboriaud C. The crystal structure of the zymogen catalytic domain of complement protease C1r reveals that a disruptive mechanical stress is required to trigger activation of the C1 complex. EMBO J. 2002;21:231–239. doi: 10.1093/emboj/21.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garnier G., Circolo A., Xu Y., Volanakis J. E. Complement C1r and C1s genes are duplicated in the mouse: differential expression generates alternative isomorphs in the liver and in the male reproductive system. Biochem. J. 2003;371:631–640. doi: 10.1042/BJ20021555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Circolo A., Garnier G., Volanakis J. E. A novel murine complement-related gene encoding a C1r-like serum protein. Mol. Immunol. 2003;39:899–906. doi: 10.1016/s0161-5890(02)00283-3. [DOI] [PubMed] [Google Scholar]
- 25a.Ligoudistianou C., Garnier G., Circolo A., Volanakis J. E. C1r-like protein (C1r-LP): a novel human complement-related protein (Abstr. XIX Int. Complement Workshop, 22–26 September 2002, Palermo, Italy) Int. Immunopharmacol. 2002;2:1267–1268. [Google Scholar]
- 26.Chirgwin J. M., Przybyla A. E., Mac Donald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 27.Steinbuch M., Audran R. The isolation of IgG from mammalian sera with the aid of caprylic acid. Arch. Biochem. Biophys. 1969;134:279–284. doi: 10.1016/0003-9861(69)90285-9. [DOI] [PubMed] [Google Scholar]
- 28.Circolo A., Nutter T. B., Strunk R. C. Biosynthesis of complement components. In: Sim R., Dodds A., editors. Complement: A Practical Approach. Oxford, U.K.: Oxford University Press; 1997. pp. 199–221. [Google Scholar]
- 29.Kim S., Narayana S. V. L., Volanakis J. E. Mutational analysis of the substrate binding site of human complement factor D. Biochemistry. 1994;33:14393–14399. doi: 10.1021/bi00252a004. [DOI] [PubMed] [Google Scholar]
- 30.Kam C. M., McRae B. J., Harper J. W., Niemann M. A., Volanakis J. E., Powers J. C. Human complement proteins D, C2, and B. Active site mapping with peptide thioester substrates. J. Biol. Chem. 1987;262:3444–3451. [PubMed] [Google Scholar]
- 31.Ponnuraj K., Xu Y., Macon K., Moore D., Volanakis J. E., Narayana S. V. L. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol. Cell. 2004;14:17–28. doi: 10.1016/s1097-2765(04)00160-1. [DOI] [PubMed] [Google Scholar]
- 32.Nakagawa M., Yuasa I., Irizawa Y., Umetsu K. The human complement component C1R gene: the exon-intron structure and the molecular basis of allelic diversity. Ann. Hum. Genet. 2003;67:207–215. doi: 10.1046/j.1469-1809.2003.00019.x. [DOI] [PubMed] [Google Scholar]
- 33.Gregory L. A., Thielens N. M., Arlaud G. J., Fontecilla-Camps J. C., Gaboriaud C. X-ray structure of the Ca2+-binding interaction domain of C1s. Insights into the assembly of the C1 complex of complement. Biol. Chem. 2003;278:32157–32164. doi: 10.1074/jbc.M305175200. [DOI] [PubMed] [Google Scholar]
- 34.Kristensen T., D'Eustachio P., Ogata R. T., Chung L. P., Reid K. B., Tack B. F. The superfamily of C3b/C4b-binding proteins. Fed. Proc. 1987;46:2463–2469. [PubMed] [Google Scholar]
- 35.Jing H., Xu Y., Carson M., Moore D., Macon K. J., Volanakis J. E., Narayana S. V. L. New structural motifs on the chymotrypsin fold and their potential roles in complement factor B. EMBO J. 2000;19:164–173. doi: 10.1093/emboj/19.2.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nonaka M., Yoshizaki F. Evolution of the complement system. Mol. Immunol. 2004;40:897–902. doi: 10.1016/j.molimm.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Khan A. R., James M. N. G. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 1998;7:815–836. doi: 10.1002/pro.5560070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patston P. A., Gettins P., Beechem J., Schapira M. Mechanism of serpin action: evidence that C1 inhibitor functions as a suicide substrate. Biochemistry. 1991;30:8876–8882. doi: 10.1021/bi00100a022. [DOI] [PubMed] [Google Scholar]
- 39.Fish W. W., Orre K., Bjork I. Routes of thrombin action in the production of proteolytically modified, secondary forms of antithrombin-thrombin complex. Eur. J. Biochem. 1979;101:39–44. doi: 10.1111/j.1432-1033.1979.tb04213.x. [DOI] [PubMed] [Google Scholar]
- 40.Ziccardi R. J., Cooper N. R. Modulation of the antigenicity of C1r and C1s by C1 inactivator. J. Immunol. 1978;121:2148–2152. [PubMed] [Google Scholar]
- 41.Bagdasarian A., Lahiri B., Talamu R. C., Wong P., Colman R. W. Immunochemical studies of plasma kallikrein. J. Clin. Invest. 1974;54:1444–1454. doi: 10.1172/JCI107892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volanakis J. E., Narayana S. V. L. Complement factor D: a novel serine protease. Protein Sci. 1996;5:553–564. doi: 10.1002/pro.5560050401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wicher K. B., Fries E. Prohaptoglobin is proteolytically cleaved in the endoplasmic reticulum by the complement C1r-like protein. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14390–14395. doi: 10.1073/pnas.0405692101. [DOI] [PMC free article] [PubMed] [Google Scholar]