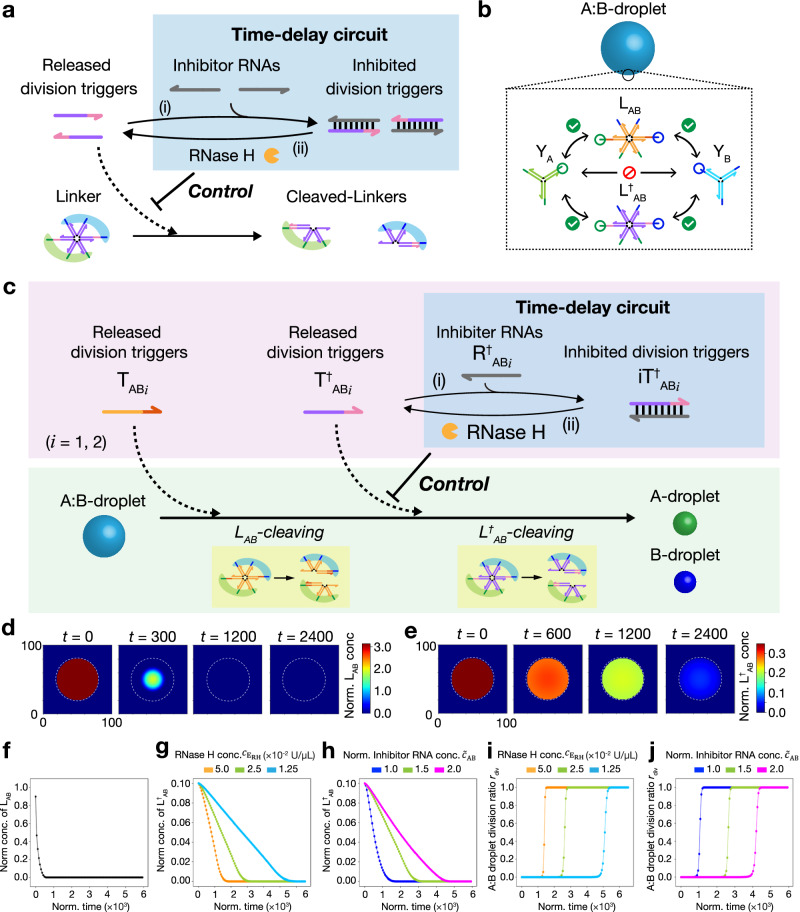
Fig. 3. Numerical investigation of timing-controlled linker-cleavage for DNA droplet division.
a Schematics of time-delay circuit to regulate cleaving rate of a DNA linker. (i) Excess inhibitor RNAs hybridize with released division triggers, producing inhibited division triggers. (ii) Released division triggers are released from inhibited division triggers by RNase H reaction. Released division triggers hybridize with the DNA linker, cleaving the linker via strand displacement. b Compositions of a binary-mixed DNA droplet (A:B-droplet). c Schematic of the timing-controlled division of A:B-droplet using a time-delay circuit. LAB is initially cleaved using TABi followed by the cleaving of L†AB, resulting in the division of A:B-droplet. The time-delayed cleaving of L†AB is achieved by the release of T†ABi from iT†ABi. The degree of time delay of the T†ABi release decides the timing control of the droplet division. Linkers and triggers with “†” indicate those that can achieve a time-delay circuit if inhibitor RNAs and RNase H are added. d, e Snapshots of numerically calculated concentrations of LAB and L†AB in the A:B-droplet using the reaction-diffusion simulation. The white broken-line circle indicates the surface of the A:B-droplet. f Time course of LAB of the A:B-droplet in the numerical simulation. g, h Time courses of L†AB concentrations of the A:B-droplet in the numerical simulation via changing the RNase H concentration or inhibitor RNA concentration , respectively. Normalized initial total concentration of inhibitor RNA is defined as = (i = 1,2), where = is the initial total concentration of excess and hybridized inhibitor RNAs, and = is the initial total concentration of released and inhibited triggers. g = 1.25, 2.5, and 5.0 × 10−2 U/µL; = 1.5. h = 2.5 × 10−2 U/µL; = 1.0, 1.5, and 2.0. i, j Time courses of the division ratio rdiv in the reaction-diffusion simulation with changing the or , respectively. i = 1.25, 2.5, and 5.0 × 10−2 U/µL; = 1.5. j = 2.5 × 10−2 U/µL; = 1.0, 1.5, and 2.0. Source data are provided as a Source Data file.