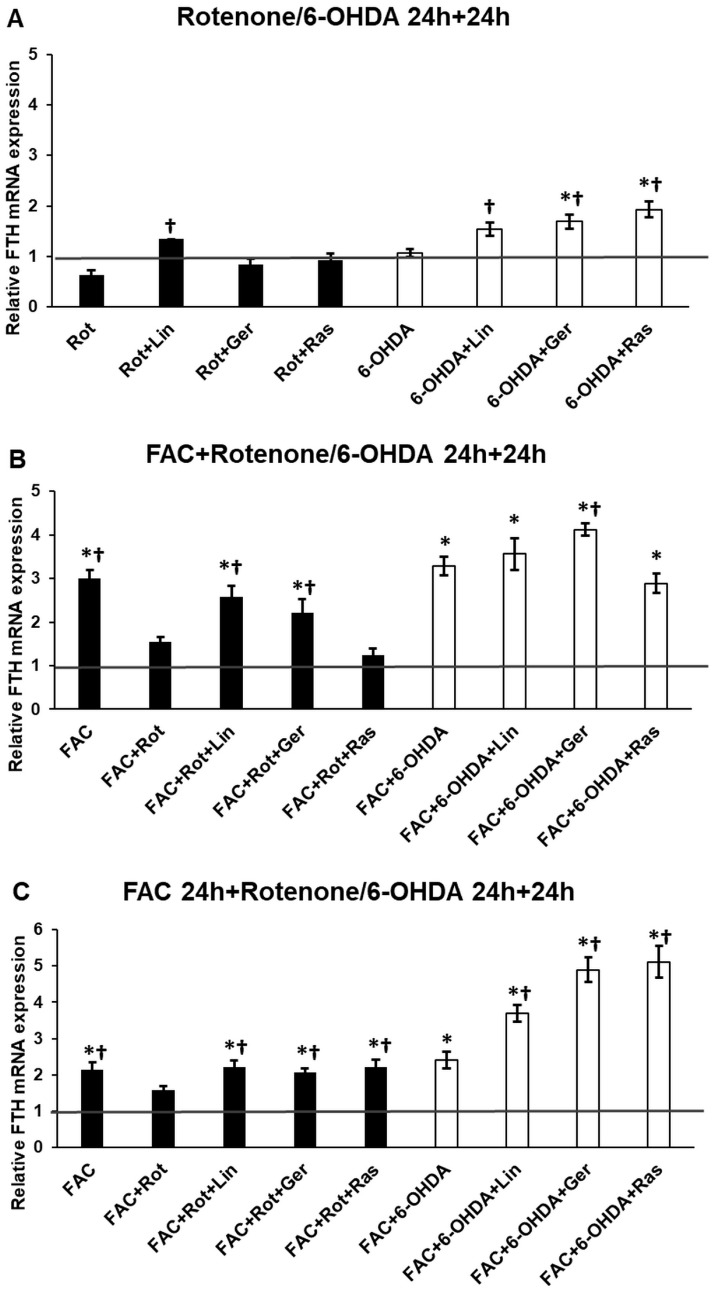
Figure 10.
Real-time PCR analysis of the FTH gene of the treated cells. The real-time PCR was performed using a SYBR Green protocol. GAPDH was used as a normalization gene. The relative mRNA expression of the control cells was considered 1 (grey horizontal line). (A) The cells were treated with rotenone or 6-OHDA for 24 h, followed by DMSO (in the case of Ctrl and Rot), linalool (Rot + Lin), geraniol (Rot + Ger), and rasagiline (Rot + Ras) for 24 h. (B) The cells were treated with ferric ammonium citrate (FAC) together with rotenone or 6-OHDA for 24 h, which was followed by DMSO (in the case of Ctrl, FAC, and FAC + Rot), linalool (FAC + Rot + Lin), geraniol (FAC + Rot + Ger), and rasagiline (FAC + Rot + Ras) for 24 h. (C) The cells were pretreated with ferric ammonium citrate (FAC) for 24 h and then were treated with rotenone or 6-OHDA for an additional 24 h, which was followed by DMSO (in the case of Ctrl, FAC, and FAC + Rot), linalool (FAC + Rot + Lin), geraniol (FAC + Rot + Ger), and rasagiline (FAC + Rot + Ras) for 24 h. The columns represent the mean value ± SD of three independent experiments. The number of technical replicates was three in each experiment. The asterisk shows p < 0.05 compared to the control. The cross indicates p < 0.05 compared to the rotenone or 6-OHDA treatments.