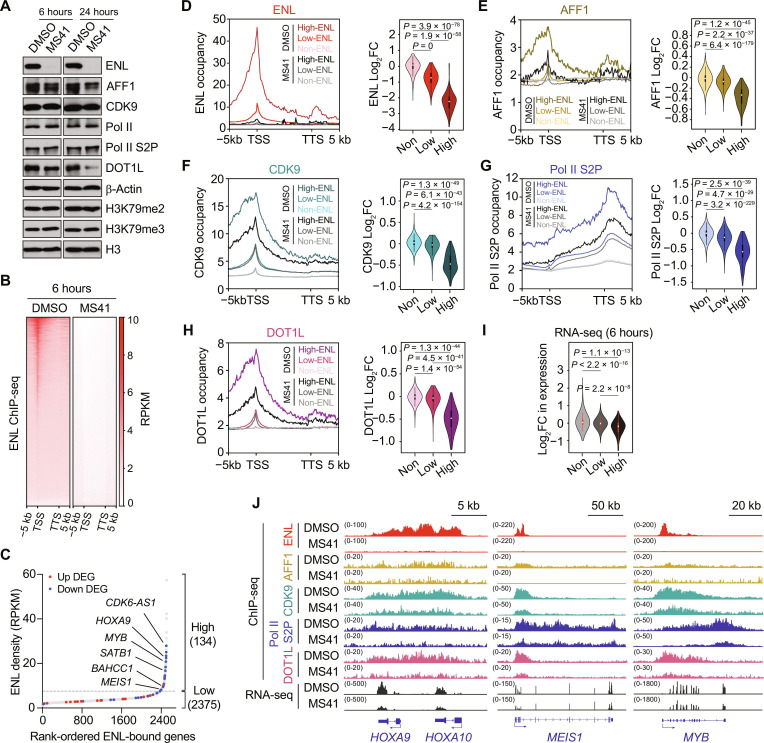
Fig. 5. MS41 reduces chromatin occupancy of ENL and its associated protein complexes.
(A) Immunoblots for ENL, AFF1, CDK9, Pol II, Pol II S2P, DOT1L, β-Actin, H3K79me2, H3K79me3, and H3 in MV4;11 cells treated with DMSO or 100 nM MS41 for 6 and 24 hours. (B) Heatmaps of ENL ChIP-seq densities on all genes in MV4;11 cells treated with DMSO or 100 nM MS41 for 6 hour. n = 3 biological replicates. RPKM, reads per kilobase million. TSS, transcription start site; TTS, transcription termination site. (C) Waterfall plot of all ENL-bound genes in 6 hours DMSO sample ranked by ENL densities from low to high in x axis. The high and low ENL–bound genes were separated on point for which a line with a slope of 1 was tangent to the curve. Red and blue dots mark up and down DEGs, respectively, in 6-hour MS41 versus DMSO-treated cells. (D to H) Average profiles of ENL (D), AFF1 (E), CDK9 (F), RNA Pol II S2P (G), and DOT1L (H) ChIP-seq densities on high-, low-, and non-ENL–bound genes as defined in (B) and (C) (left). Violin plots of log2 fold changes (log2 FC) of corresponding ChIP-seq densities in MS41 versus DMSO from the left panel (right). Two-tailed t test was used for calculating P values. (I) Violin plot of log2 FC of 6-hour MS41 versus DMSO RNA-seq CPM values. Two-tailed t test was used for calculating P values. (J) Integrative Genomics Viewer (IGV) views of ChIP-seq densities of (D) to (H) and RNA-seq reads on HOXA9/10, MEIS1, and MYB genes.