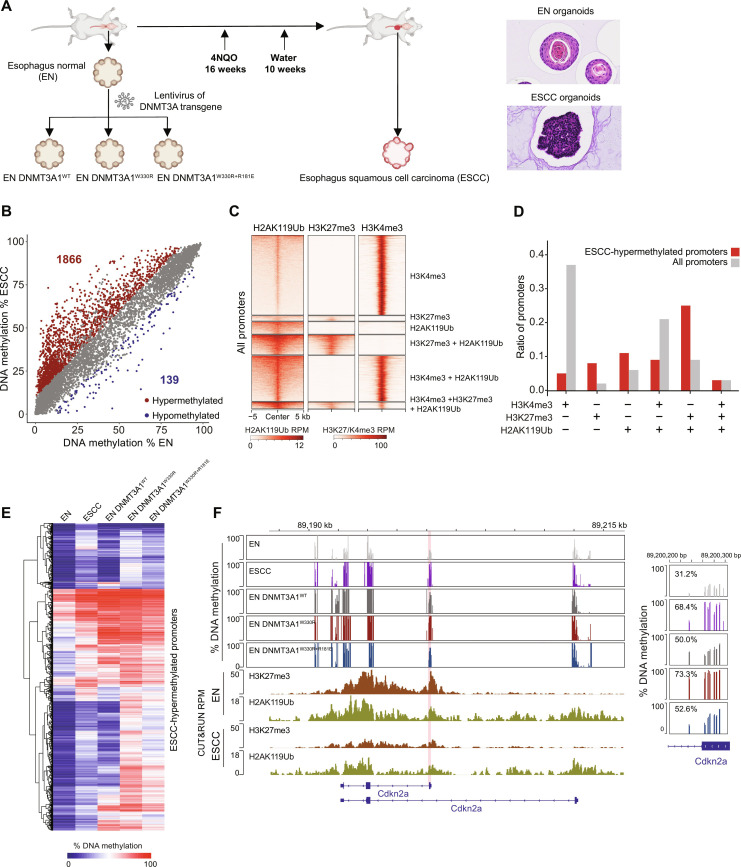
Fig. 6. Cancer-associated Polycomb CGI hypermethylation requires DNMT3A1–acidic patch interaction.
(A) Schematic of EN- and 4NQO-induced ESCC organoid collection. Right: Representative images of EN and ESCC organoid histology. (B) Scatter plots showing the percentage of DNAme (by RRBS) at all promoters (TSS ± 500 bp) in ESCC versus EN organoids. Each dot represents a single promoter. Significantly hypermethylated (>10% and q < 0.01) and hypomethylated (<−10% and q < 0.01) promoters are respectively colored in red and blue (three biological replicates), respectively. (C) Heatmap showing enrichment of H3K27me3, H2AK119Ub, and H3K4me3 CUT&Tag reads at all promoters (TSS ± 5 kb) in EN organoids. Promoters are classified to six groups: H3K4me3+; H3K27me3+; H2AK119Ub+; H3K4me3+ and H2AK119Ub+; H3K27me3+ and H2AK119Ub+; and H3K4me3+, H3K27me3+, and H2AK119Ub+. (D) Bar plots showing the representation among all promoters, or ESCC-hypermethylated promoters, for each promoter group defined in (C). (E) Heatmap showing the percentage of promoter DNAme (by RRBS) in EN organoids, ESCC organoids, and EN organoids expressing DNMT3A1WT, DNMT3A1W330R, and DNMT3A1W330R+R181E for all ESCC-hypermethylated promoters. (F) Genome browser view of Cdkn2a (chromosome 4, 89,187 to 89,216 Mb, GRCm39), showing the percentage of DNAme (by RRBS) in EN organoids, ESCC organoids, and EN organoids expressing DNMT3A1WT, DNMT3A1W330R, and DNMT3A1W330R+R181E. Bottom: H3K27me3 and H2AK119Ub CUT&Tag reads in EN and ESCC organoids. Right: Average DNAme at Cdkn2a promoter (chromosome 4, 89,200,200 to 89,200,300 bp, GRCm39). Genes from RefSeq database are annotated at the bottom.