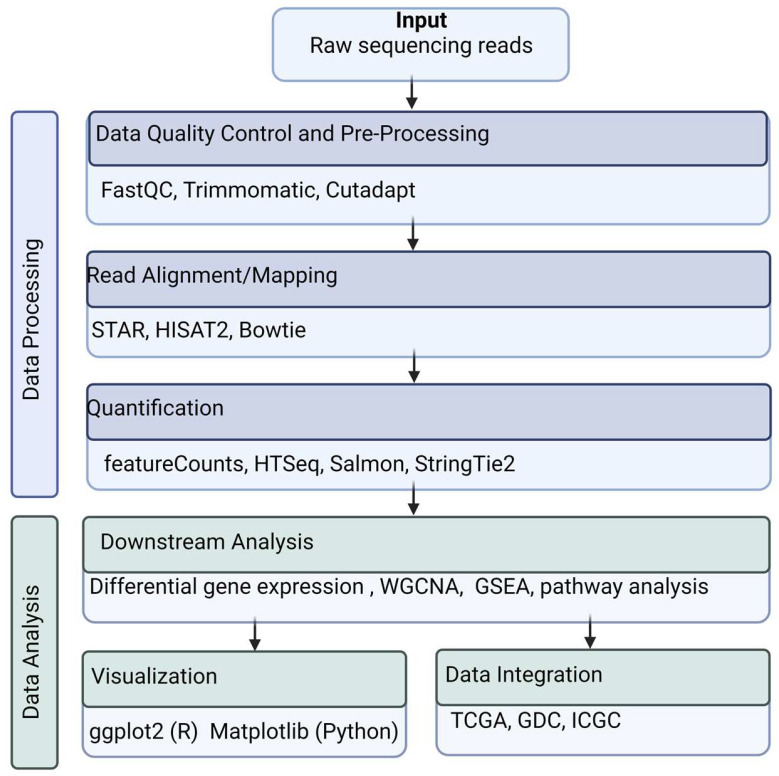
Figure 2.
Overview of RNA-Seq data workflow. In this example of RNA-Seq data analysis workflow, raw sequence reads serve as the input. Raw sequences undergo pre-processing and quality control. Next, the Fastq files are aligned to a reference genome (SAM/BAM files) and are quantified to generate count matrices (Text files). Downstream analysis, such as differential gene expression, uses the count matrices as input. The data results can be visualized and integrated using R packages, Python libraries, and other tools.