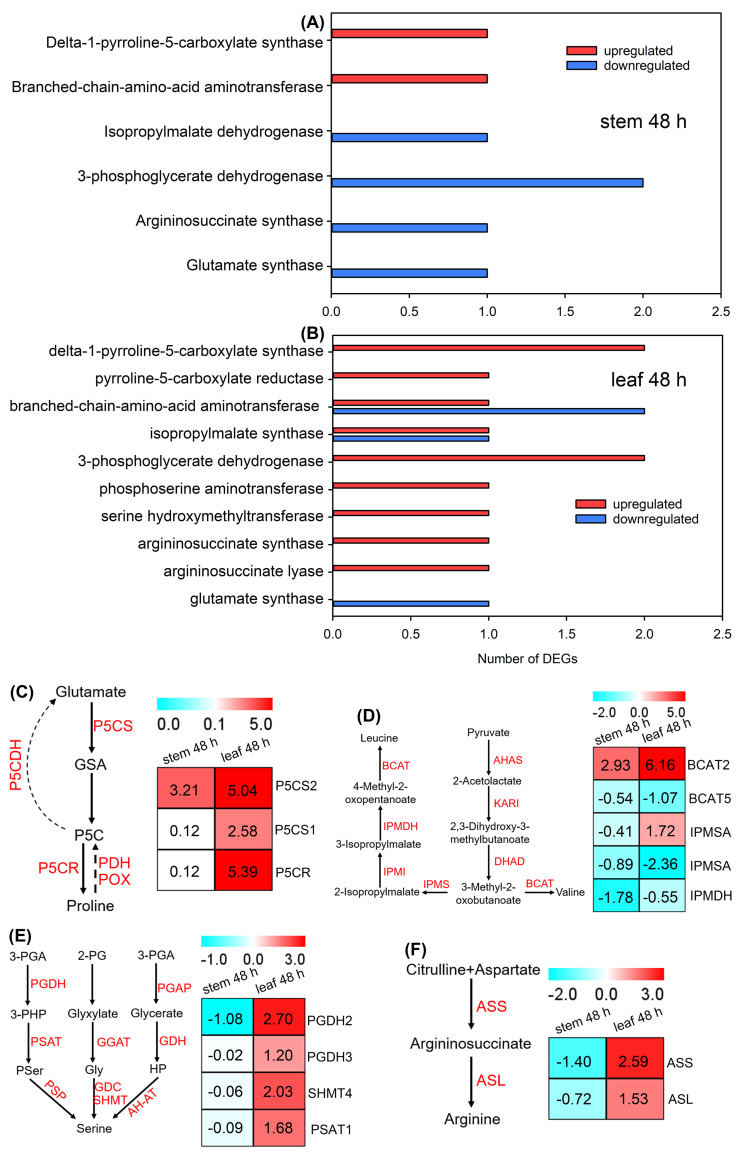
Figure 8.
The analysis of DEGs related to the amino acid metabolism in sweet sorghum after 20% PEG treatment. (A,B) The number of DEGs identified in the stems and leaves after PEG treatment for 48 h, respectively; (C) the expression level of genes related to proline metabolism; (D) the expression level of genes related to leucine and valine biosynthesis; (E) the expression level of genes related to serine biosynthesis; (F) the expression level of genes related to arginine biosynthesis. The number in each block refers to the fold changes in each metabolite. GSA, glutamate-semialdehyde; P5C, pyrroline-5-carboxylic acid; P5CS, delta-1-pyrroline-5-carboxylate synthase; P5CR, pyrroline-5-carboxylate reductase; PDH, proline dehydrogenase; POX, proline oxidase; P5CDH, pyrroline-5-carboxylic acid dehydrogenase; AHAS, acetohydroxyacid synthase; KARI, ketolacid reducto-isomerase; DHAD, dihydroxyacid dehydratase; BCAT, branched-chain aminotransferase; IPMS, isopropylmalate synthase; IPMI, isopropylmalate isomerase; IPMDH, isopropylmalate dehydrogenase; 3-PGA, 3-phosphoglycerate; 2-PG, 2-phosphoglycolate; PGDH, phosphoglycerate dehydrogenase; PGAP, 3-PGA phosphatase; PSAT, phosphoserine aminotransferase; GGAT, glyoxylate glutamate aminotransferase; GDH, glycerate dehydrogenase; PSP, phosphoserine phosphatase; GDC, glycine decarboxylase complex; SHMT, serine hydroxymethyltransferase; AH-AT, alanine-HP aminotransferase; ASS, argininosuccinate synthase; ASL, argininosuccinate lyase.