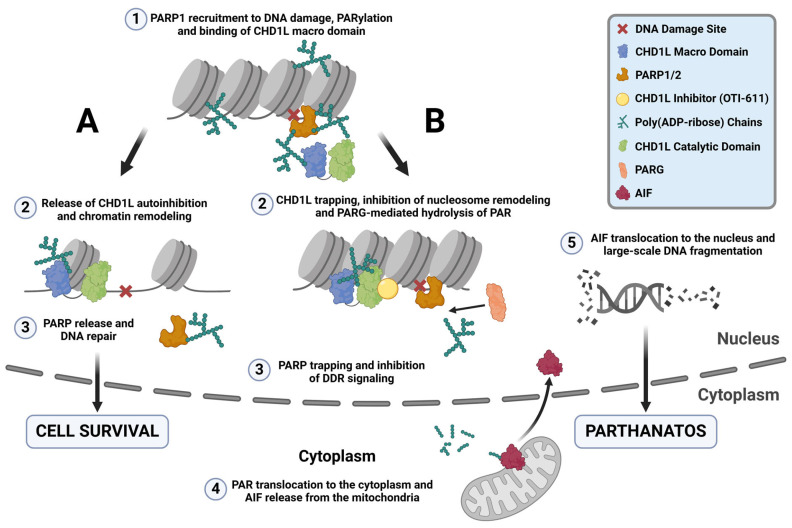
Figure 8.
CHD1L ATPase inhibition traps CHD1L at DNA damage sites and induces PARthanatos. (A) Under normal conditions, upon DNA damage, PARP1/2 detect SSB and DSB and recruit repair machinery to damage sites by auto-PARylating themselves and through PARylating other repair proteins, and histones. One of these proteins is CHD1L, which binds to the PAR chains by its macro domain releasing its autoinhibition. Once CHD1L is activated, it can bind the histone and relax the chromatin through its ATPase-driven chromatin remodeling activity, promoting DNA repair and cell survival. PARP1 is released from the DNA damage site and PAR is recycled through PARG-mediated hydrolysis. (B) When CHD1L ATPase is inhibited by OTI-611, CHD1L becomes trapped near DNA damage sites and unable to relax the chromatin. Moreover, unprotection of the PAR chains causes their PARG-mediated hydrolysis, trapping PARP1/2. This mechanism of PARP trapping does not cause DNA damage, unlike PARPi entrapment of PARP on relaxed chromatin, which undergoes DNA repair and subsequent replication fork collapse in HR-deficient tumor cells. Additionally, CHD1L inhibition by OTI-611 leaves PAR chains unprotected, allowing PARG to hydrolyze PAR and enable PAR fragment translocation to the cytoplasm. In the cytoplasm, AIF binds to PAR fragments in the mitochondria, causing its release and subsequent translocation to the nucleus. Once in the nucleus, AIF triggers large-scale DNA fragmentation and a form of non-apoptotic cell death known as PARthanatos.