Abstract
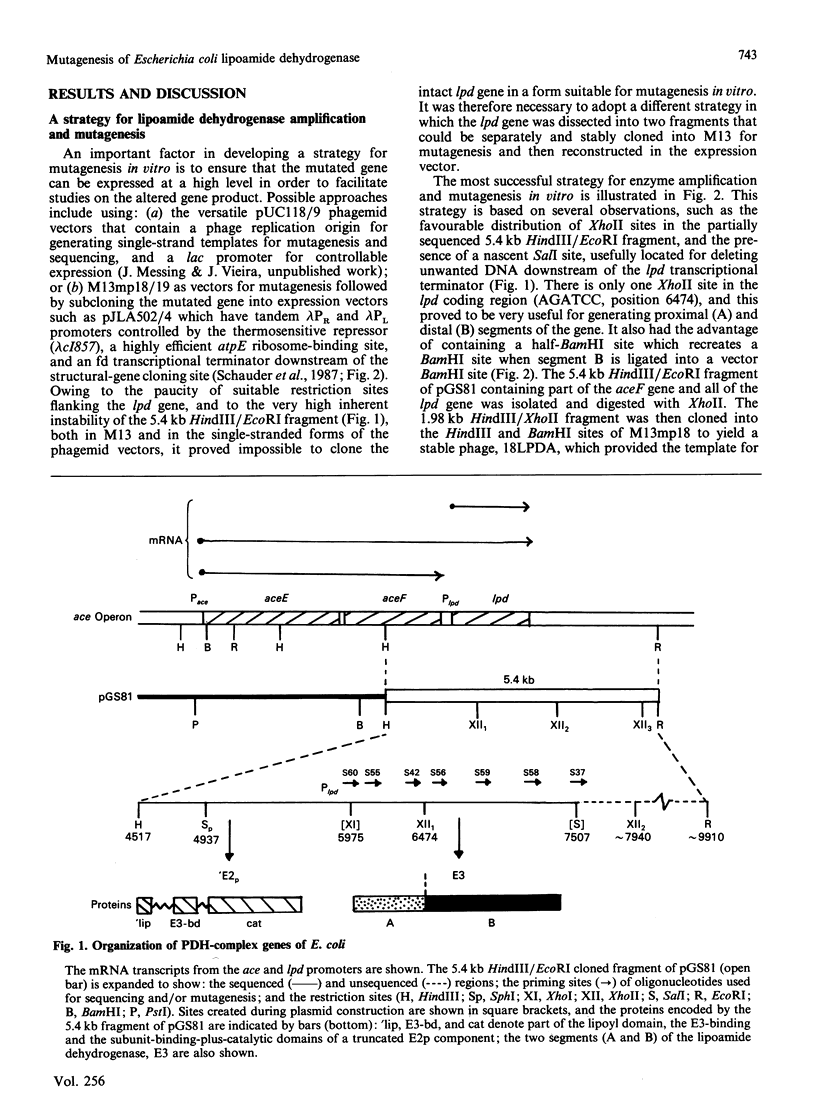
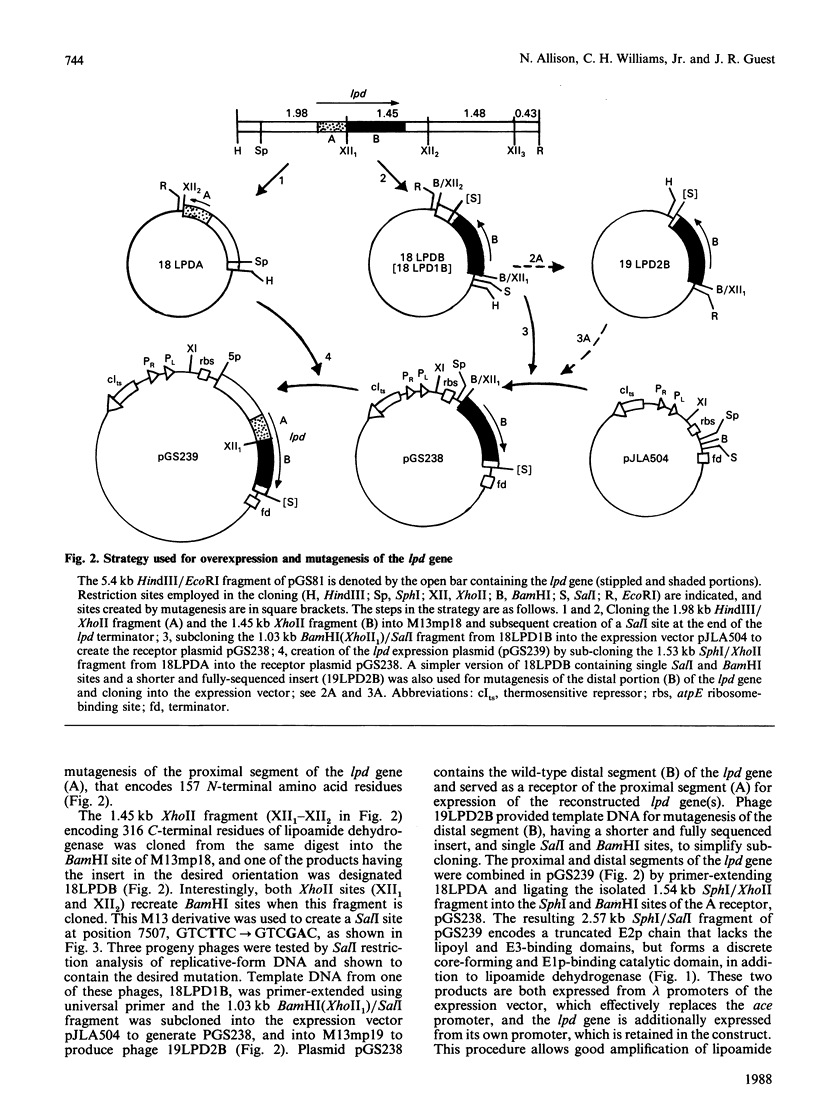
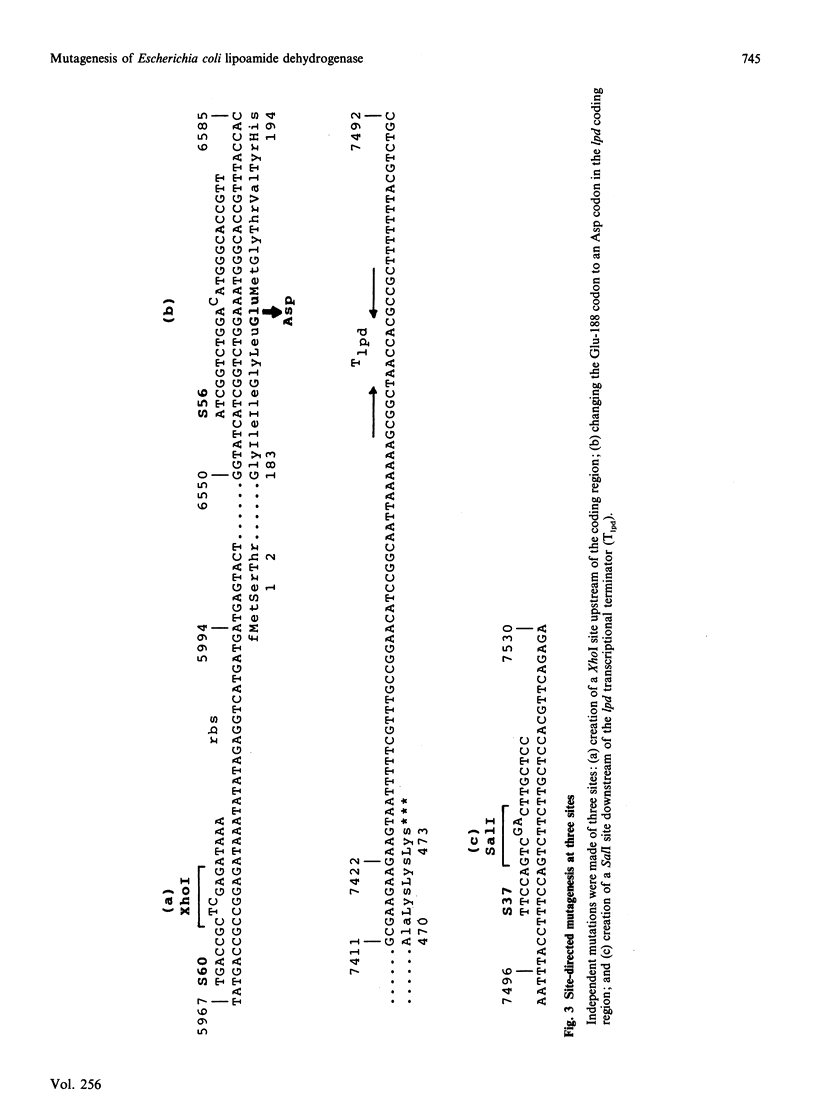
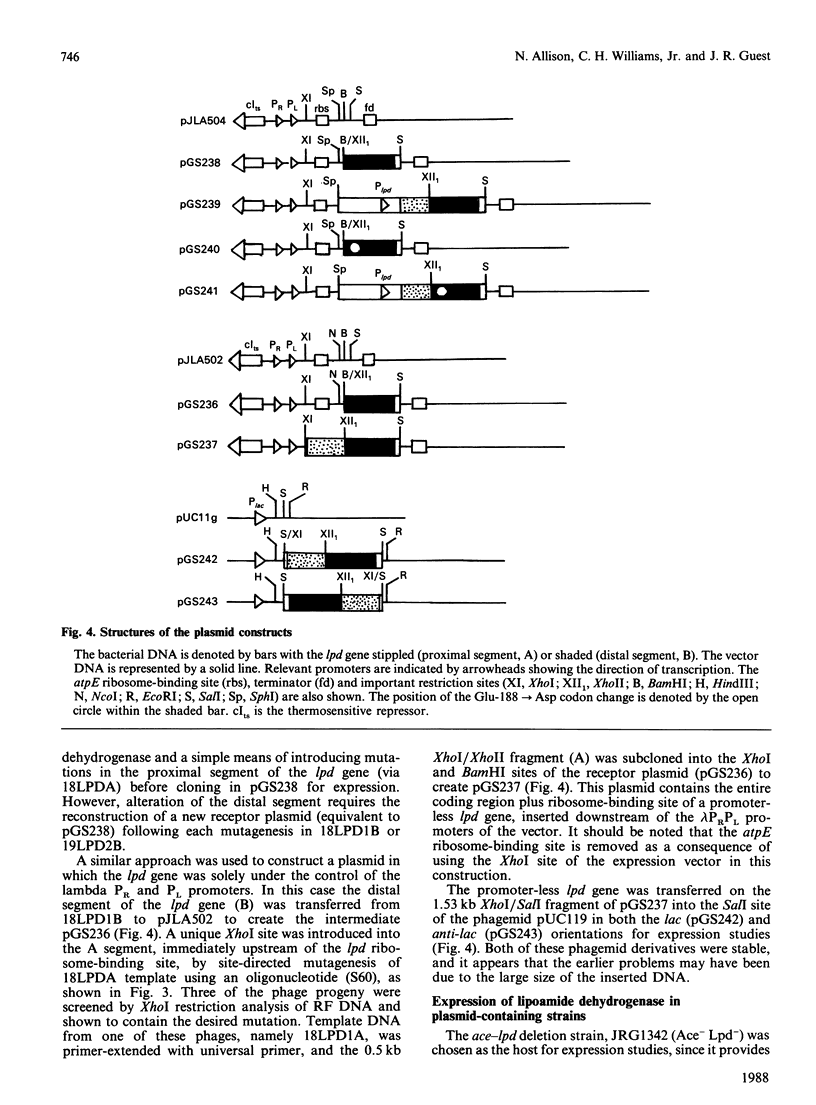
A 'split-gene' technique for the overexpression and mutagenesis of the gene encoding the lipoamide dehydrogenase of Escherichia coli was developed in order to overcome the instability problems encountered when attempting to mutate the intact gene. The lipoamide dehydrogenase gene, lpd, was dissected into two fragments which were separately subcloned into M13 vectors for mutagenesis in vitro followed by reconstitution in the pJLA504 expression vector under the transcriptional control of the lambda PR and lambda PL promoters and a temperature-sensitive lambda repressor. After thermo-induction, E. coli cells transformed with the plasmid carrying the reconstituted lpd gene contained 4-5 times more lipoamide dehydrogenase activity than is normally found in the wild-type organism. The strategy was used to engineer a Glu-188----Asp replacement in lipoamide dehydrogenase, and this generated an enzyme with markedly different kinetic properties.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson M. J., Conroy K., McQuattie A., Stevenson K. J. Dihydrolipoamide dehydrogenase from Trypanosoma brucei. Characterization and cellular location. Biochem J. 1987 May 1;243(3):661–665. doi: 10.1042/bj2430661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox B., Walsh C. T. Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J Biol Chem. 1982 Mar 10;257(5):2498–2503. [PubMed] [Google Scholar]
- Guest J. R. Aspects of the molecular biology of lipoamide dehydrogenase. Adv Neurol. 1978;21:219–244. [PubMed] [Google Scholar]
- Guest J. R., Lewis H. M., Graham L. D., Packman L. C., Perham R. N. Genetic reconstruction and functional analysis of the repeating lipoyl domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Mol Biol. 1985 Oct 20;185(4):743–754. doi: 10.1016/0022-2836(85)90059-2. [DOI] [PubMed] [Google Scholar]
- Guest J. R., Roberts R. E., Stephens P. E. Hybrid plasmids containing the pyruvate dehydrogenase complex genes and gene-DNA relationships in the 2 to 3 minute region of the Escherichia coli chromosome. J Gen Microbiol. 1983 Mar;129(3):671–680. doi: 10.1099/00221287-129-3-671. [DOI] [PubMed] [Google Scholar]
- Guest J. R., Stephens P. E. Molecular cloning of the pyruvate dehydrogenase complex genes of Escherichia coli. J Gen Microbiol. 1980 Dec;121(2):277–292. doi: 10.1099/00221287-121-2-277. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Pyridine nucleotide - disulfide oxidoreductases. Experientia Suppl. 1980;36:149–180. doi: 10.1007/978-3-0348-5419-1_5. [DOI] [PubMed] [Google Scholar]
- Kochi H., Kikuchi G. Mechanism of reversible glycine cleavage reaction in Arthrobacter globiformis. Function of lipoic acid in the cleavage and synthesis of blycine. Arch Biochem Biophys. 1976 Mar;173(1):71–81. doi: 10.1016/0003-9861(76)90236-8. [DOI] [PubMed] [Google Scholar]
- Krauth-Siegel R. L., Blatterspiel R., Saleh M., Schiltz E., Schirmer R. H., Untucht-Grau R. Glutathione reductase from human erythrocytes. The sequences of the NADPH domain and of the interface domain. Eur J Biochem. 1982 Jan;121(2):259–267. doi: 10.1111/j.1432-1033.1982.tb05780.x. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langley D., Guest J. R. Biochemical genetics of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: isolation and biochemical properties of deletion mutants. J Gen Microbiol. 1977 Apr;99(2):263–276. doi: 10.1099/00221287-99-2-263. [DOI] [PubMed] [Google Scholar]
- McCully V., Burns G., Sokatch J. R. Resolution of branched-chain oxo acid dehydrogenase complex of Pseudomonas aeruginosa PAO. Biochem J. 1986 Feb 1;233(3):737–742. doi: 10.1042/bj2330737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J. S., Guest J. R. Subgenes expressing single lipoyl domains of the pyruvate dehydrogenase complex of Escherichia coli. Biochem J. 1987 Aug 1;245(3):869–874. doi: 10.1042/bj2450869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai E. F., Schulz G. E. The catalytic mechanism of glutathione reductase as derived from x-ray diffraction analyses of reaction intermediates. J Biol Chem. 1983 Feb 10;258(3):1752–1757. [PubMed] [Google Scholar]
- Rice D. W., Schulz G. E., Guest J. R. Structural relationship between glutathione reductase and lipoamide dehydrogenase. J Mol Biol. 1984 Apr 15;174(3):483–496. doi: 10.1016/0022-2836(84)90332-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schauder B., Blöcker H., Frank R., McCarthy J. E. Inducible expression vectors incorporating the Escherichia coli atpE translational initiation region. Gene. 1987;52(2-3):279–283. doi: 10.1016/0378-1119(87)90054-0. [DOI] [PubMed] [Google Scholar]
- Schulz G. E., Schirmer R. H., Sachsenheimer W., Pai E. F. The structure of the flavoenzyme glutathione reductase. Nature. 1978 May 11;273(5658):120–124. doi: 10.1038/273120a0. [DOI] [PubMed] [Google Scholar]
- Spencer M. E., Guest J. R. Transcription analysis of the sucAB, aceEF and lpd genes of Escherichia coli. Mol Gen Genet. 1985;200(1):145–154. doi: 10.1007/BF00383328. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Lewis H. M., Darlison M. G., Guest J. R. Nucleotide sequence of the lipoamide dehydrogenase gene of Escherichia coli K12. Eur J Biochem. 1983 Oct 3;135(3):519–527. doi: 10.1111/j.1432-1033.1983.tb07683.x. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme R., Pai E. F., Schirmer R. H., Schulz G. E. Three-dimensional structure of glutathione reductase at 2 A resolution. J Mol Biol. 1981 Nov 15;152(4):763–782. doi: 10.1016/0022-2836(81)90126-1. [DOI] [PubMed] [Google Scholar]
- Williams C. H., Jr, Arscott L. D., Schulz G. E. Amino acid sequence homology between pig heart lipoamide dehydrogenase and human erythrocyte glutathione reductase. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2199–2201. doi: 10.1073/pnas.79.7.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]



