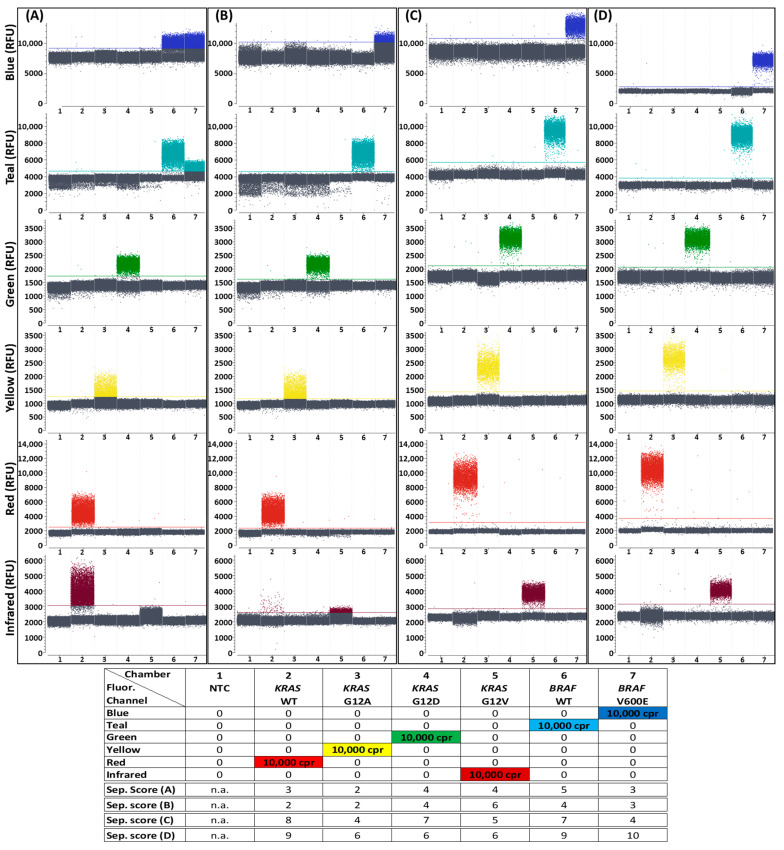
Figure 6.
Improving discriminability between positive and negative signal populations in dPCR reactions step-by-step. During all steps of generic reporter set optimization, the target panel for detection of KRAS and BRAF mutations and their corresponding WT controls was used. 1 D-plots of all 6 fluorescence channels are shown. Reaction chamber details from left to right: (1) NTC; (2–7) 10,000 cpr of one kind of target sequence each. A more detailed version of this figure, with samples containing all six targets at once, is shown in Figure S5. Separability scores between positive and negative droplet populations are indicated at the bottom of the legend. Fluorophores for this second generic reporter set were selected according to the MEA data shown in Figure S3. Details of the used reporters in each detection channel including sequences, fluorophores and quenchers can be seen in Table S2. (A) Initial non-optimized test without color compensation. (B) Initial non-optimized test after setting a suitable color compensation, with which each target is detected in only one fluorescence channel. This color compensation matrix was henceforth used in every subsequent test to optimize the second generic reporter set. (C) Improved separability after increasing the PCR cycle number from 45 to 60. (D) Further improved separability after reduction of background fluorescence in the “Blue” and “Teal” channels by changing the naica® master mix version from “naica® multiplex PCR MIX” to “naica® PCR MIX”. Although the second generic reporter set shown here is an optimized set, ready to be used in combination with other target panels, it utilizes different reporters from the first 6-plex generic reporter set that was characterized in the KRAS, BRAF and NRAS quantification studies shown in Section 2.1.