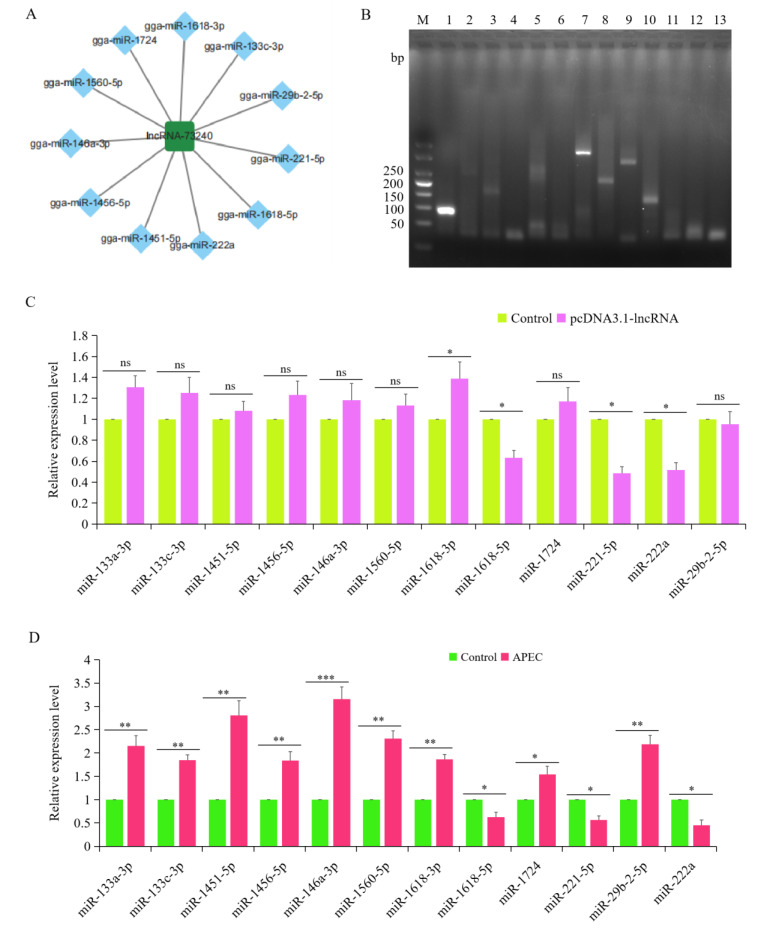
Figure 2.
Construction of lincRNA-73240–miRNA–mRNA networks. (A) Potential miRNAs that interact with lincRNA-73240. (B) Agarose gel electrophoresis was used to identify RT-qPCR amplification products of potential miRNAs: 1, U6; 2, miR-133a-3p; 3, miR-133c-3p; 4, miR-1451-5p; 5, miR-1456-5p; 6, miR-146a-3p; 7, miR-1560-5p; 8, miR-1618-3p; 9, miR-1618-5p; 10, miR-1724; 11, miR-221-5p; 11, miR-222a; 12, gga-miR-29b-2-5p. (C) Expression levels of the potential miRNAs during APEC infection. Data expressed as mean ± SD of four independent experiments; each experiment performed in triplicate; ANOVA test; ns, not significant; * p < 0.05. (D) Effects of lincRNA-73240 on the potential miRNAs that interact with lincRNA-73240. Data are shown as means ± SD; n = 4 independent experiments; each experiment performed in triplicate; ANOVA test; ns, not significant; * p < 0.05; ** p < 0.01; *** p < 0.001.