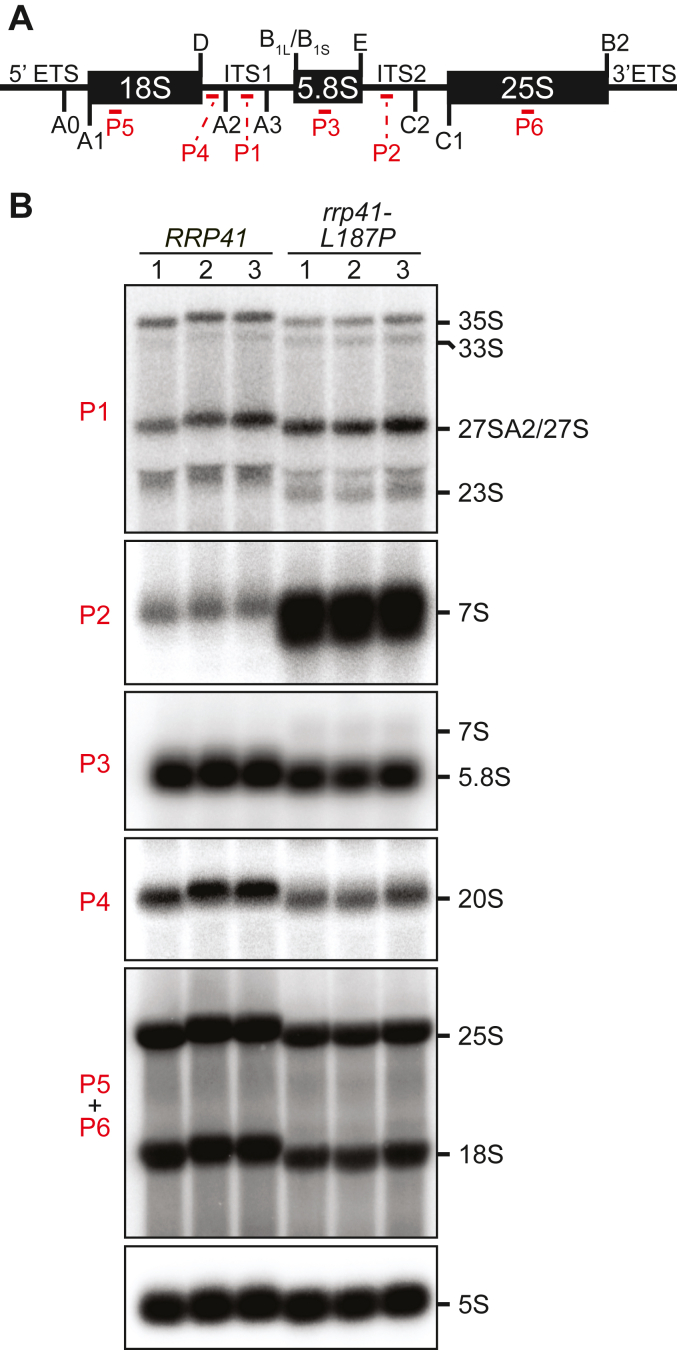
Figure 5.
The rrp41-L187P cells exhibit defects in rRNA processing. A, schematic of yeast 35S rRNA indicating the locations of rRNA cleavage sites and northern blot probes used (P1-P6; red). The 35S rRNA transcript contains 18S, 5.8S, and 25S rRNA separated by internal transcribed spacer 1 and 2 (ITS1, ITS2) and flanked by 5′ and 3′ external transcribed spacer (5′ETS, 3′ETS). B, northern blotting of three independent biological replicates of rrp41-L187P CRISPR mutant and WT (RRP41) cells reveals rRNA processing defects in rrp41-L187P mutant cells, including the accumulation of 7S pre-rRNA. Equal amounts of total RNA from control WT RRP41 cells or rrp41-L187P cells grown at 30 °C was used for northern blotting. The 5S probe was used a loading control.