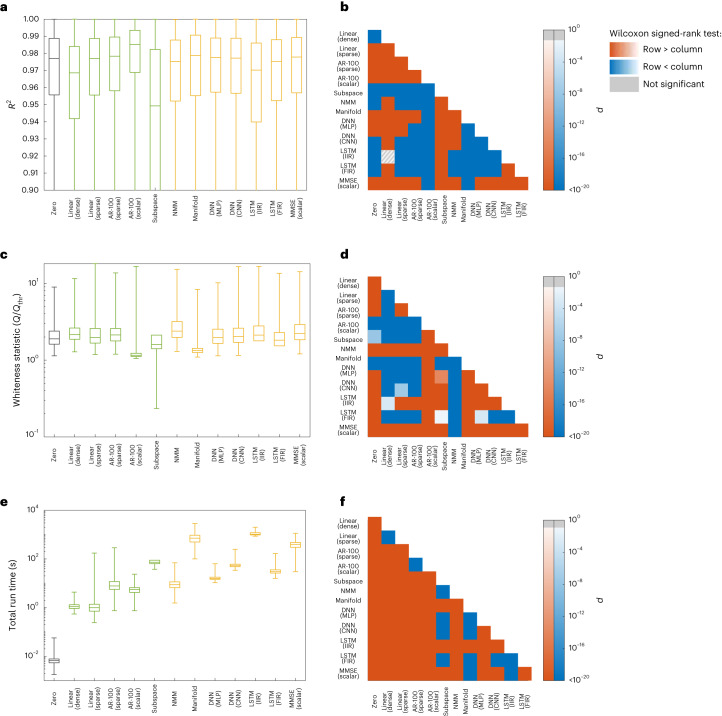
Fig. 3. Linear vs nonlinear models of rsiEEG activity.
Panels and acronyms parallel those in Fig. 2. a, The distribution of cross-validated regional , combined across all electrodes (the number of which varies among participants) and all the recording sessions of the 122 participants (sample size = 776,484). Linear and nonlinear methods are depicted by green and yellow boxes, respectively (see Methods for an explanation of each model). Unlike data presented in Fig. 2, pairwise linear or pairwise MMSE models are not included due to the observation that between-electrode connections decrease the cross-validated accuracy of the top model (cf. the 4th and 5th boxplots). In contrast, including scalar autoregressive lags is highly beneficial in iEEG, whereas it is not so in rsfMRI. Therefore, the MMSE model here is scalar, conditioning on the past lags of each region itself. The lower whisker of the boxplots are trimmed to allow for better illustration of the interquartile ranges. b, The P value of the one-sided Wilcoxon signed-rank test performed between all pairs of distributions of R2 in a. Warm (cold) colours indicate that the distribution labelled on the row is significantly larger (smaller) than the distribution labelled on the column. Grey hatches indicate non-significant differences evaluated at α = 0.05 with BH-FDR correction for multiple comparisons. c,d, Similar to a and b but for the statistic Q of the multivariate test of whiteness relative to its rejection threshold Qthr (cf. Methods). Smaller Q/Qthr indicates whiter (better) residuals, with Q/Qthr ≤ 1 required for the null hypothesis of whiteness not to be rejected. e,f, Similar to a and b but for the time that it took for the learning and out-of-sample prediction of each model. In all boxplots, the centre line, box limits and whiskers represent the median, upper and lower quartiles, and the smallest and largest samples, respectively.