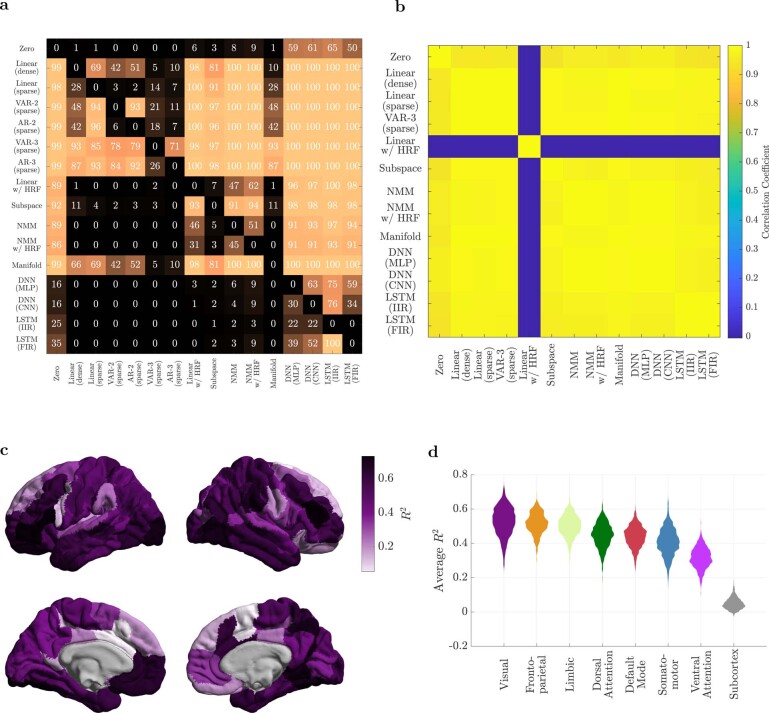
Extended Data Fig. 4. The spatial distribution of predictability in fMRI data.
(a) Regionwise comparisons between all pairs of models. The color and number in each cell indicates the percentage of brain regions where the median of the R2 values of the model on the row is statistically significantly greater that the median of the R2 values of the model on the column (α = 0.05, Wilcoxon signed rank test, and Bonferroni correction for multiple comparisons). (b) The correlation coefficient between the R2 values of all the (brain-wise) methods. All methods produce almost the same cortical distributions of R2 (albeit with different absolute values of R2, cf. Fig. 2a in the main text), except for ‘Linear w/ HRF’ which has an un-correlated R2 distribution relative to the rest of the methods. (c) The cortical distribution of the R2 of our best model (‘VAR-3 (sparse)’), averaged over the 700 subjects. (d) Violin plots of the distribution of R2, averaged over all the regions of each resting state network, for all subjects. Note that each distribution in panel (b) is thus composed of 700 samples. All pairs of distributions have significantly different medians in the order plotted (one-sided Mann-Whitney U-test at α = 0.05 with BH-FDR correction for multiple comparisons). The most striking difference is between the cortical and subcortical regions, where the dynamics of the latter are remarkably less predictable than the former. This lower predictability can be viewed as having ‘more noisy’ dynamics or, more precisely, less spatially and temporally correlated (that is, more white) fMRI time series in these regions.