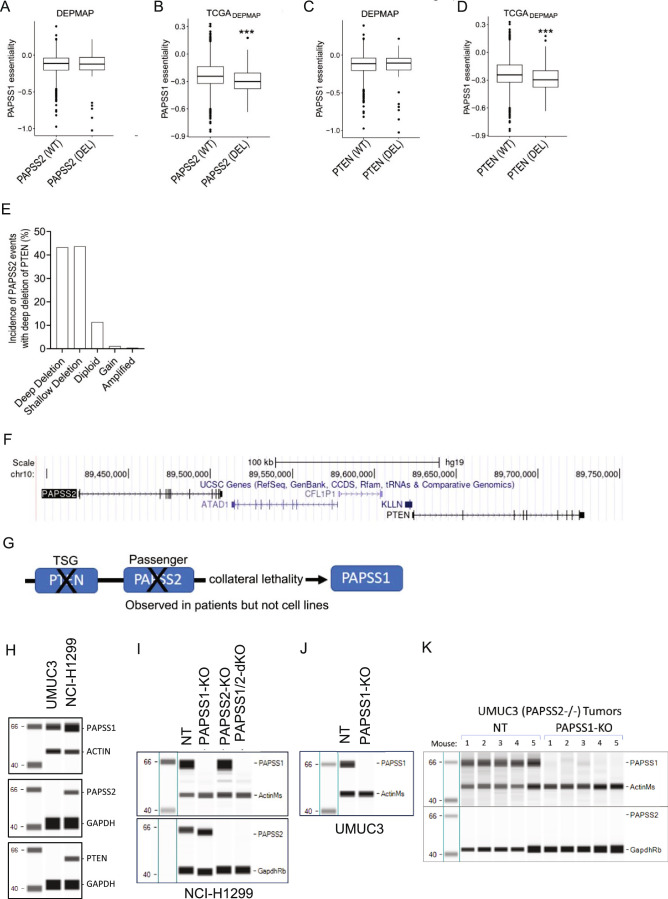
Extended Data Fig. 4. Supporting evidence of PAPSS1/2 synthetic lethality.
a, b) PAPSS1 is a novel synthetic lethality in the context of PAPSS2 deletion, which is not detectable in (a) DEPMAP cell lines (n = 905) and is only detectable in (b) TCGADEPMAP patient samples (n = 7,581). (c, d) Likewise, PAPSS1 is not synthetic lethal with PTEN deletion in DEPMAP cell lines (c, n = 905) and is only detectable in TCGADEPMAP patient samples (d, n = 7,581). For (A-D), the center horizontal line represents the median (50th percentile) value. The box spans from the 25th to the 75th percentile. The whiskers indicate the 5th and 95th percentiles. (e) Unlike cultured cell models, PAPSS2 is frequently co-deleted with PTEN in TCGA patients. (f) PAPSS2 is a closely neighboring gene of PTEN. (g) A schematic representation summarizing the hypothesized synthetic lethality of PAPSS1 that is driven by collateral deletion of PAPSS2 with the tumor suppressor gene (TSG), PTEN, in patients but not cell lines. ***P < 0.001, as determined by the Wilcoxon rank-sum test. (h) Endogenous expression by Simple Western of PAPSS1, PAPSS2, and PTEN in the model cell lines UMUC3 and NCI-H1299. (i,j) Validation of PAPSS1 and PAPSS2 single (KO) and double (dKO) knockouts by RNP in spheroid experiments for NCI-H1299 (i) and UMUC3 (j). (k) Validation of PAPSS1 knockout in the UMUC3 xenograft experiment tumors (n = 5 tumors per condition from n = 1 independent experiment). Molecular weight marker lanes are shown in kDa. Data shown in (h-j) are representative from at least 3 independent experiments. The two-sided Wilcoxon rank test was used for (A-D), ***P < 0.001.