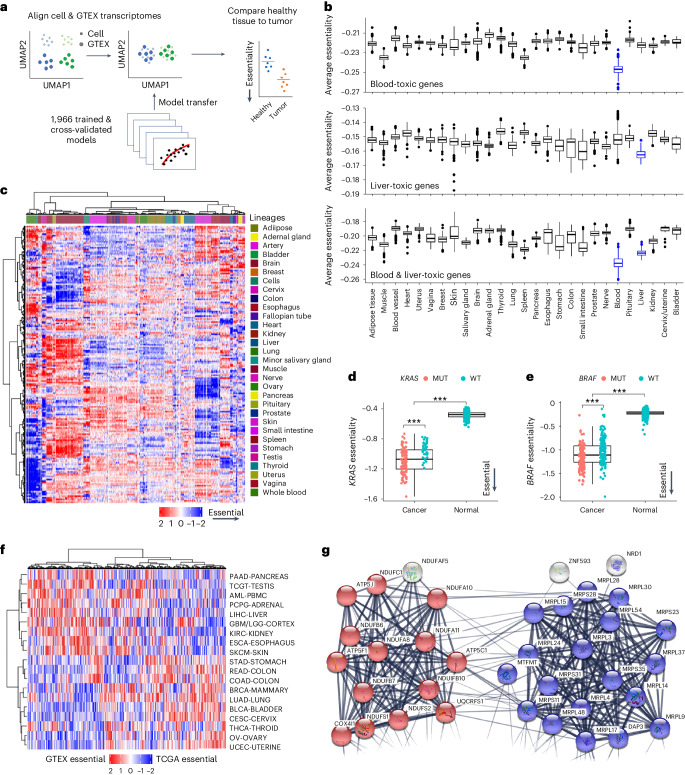
Fig. 7. Building a translational dependency map in normal tissues: GTEXDEPMAP.
a, Schematic of gene essentiality model transposition from DEPMAP to GTEX, following alignment of genome-wide expression data to account for differences in homogeneous cultured cell lines and healthy tissue biopsies. b, Average gene essentiality profile across healthy tissues of GTEXDEPMAP (n = 17,382) for molecular targets with known liver and blood toxicities (in blue). c, Unsupervised clustering of predicted gene essentiality scores across healthy tissues. Blue indicates genes with stronger essentiality and red indicates genes with less essentiality. d, KRAS essentiality is significantly higher in PAAD with GOF mutations compared to healthy pancreas in GTEXDEPMAP (n = 146 for cancer with n = 106 KRASmut and n = 40 KRASwt, n = 328 for normal) e, BRAF essentiality is significantly higher in SKCM with GOF mutations compared to normal skin GTEXDEPMAP (n = 319 for cancer with n = 165 BRAFmut and n = 154 BRAFwt, n = 1,809 for normal) For b, d, and e, the center horizontal line represents the median (50th percentile) value. The box spans from the 25th to the 75th percentile. The whiskers indicate the fifth and 95th percentiles. f, Global differences between the predicted target efficacy score (TCGADEPMAP) and the healthy tissue-of-origin tolerability score (GTEXDEPMAP). g, STRING network analysis of the top 100 LUAD targets with the greatest predicted tolerability in healthy lung reveals significant connectivity (P < 1 × 10−16) and gene ontology enrichment oxidative phosphorylation (blue-colored spheres; P = 5.8 × 10−11) and mitochondrial translation (red-colored spheres; P = 2.9 × 10−20). ***P < 0.001, as determined by a Wilcoxon rank-sum test for two-group comparison and Kruskal–Wallis test followed by a Wilcoxon rank-sum test with multiple test correction for a multi-group comparison (d and e).