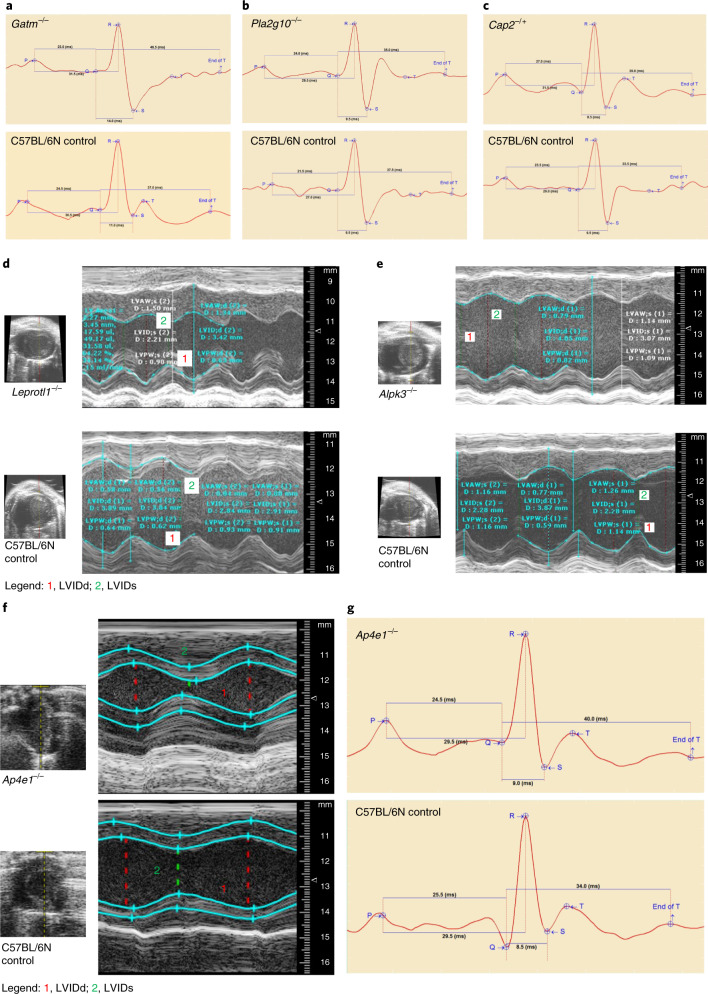
Fig. 1. Candidate genes for cardiac conduction system disease and cardiomyopathy in mice.
Representative electrocardiograms from conscious mutant and control mice with indication of ECG parameters and interval durations. a, Gatm loss (Gatm−/−) caused lower heart rate with prolonged QRS width and QTc and ST interval lengths. b, Pla2g10 loss (Pla2g10−/−) lowered heart rate and prolonged PR, PQ and QTc intervals in female null mice. c, Cap2 depletion (Cap2−/+) induced lower heart rate and lengthy QTc and ST durations in male null mice compared with C57BL/6N controls. Data are presented by Mouse Specifics software. Interval durations are given in milliseconds. M-mode recordings are through a short-axis view tangential to the papillary muscle from representative mutant and control mice. Images show the LVID throughout diastole and systole. d, Leprotl1 depletion (Leprotl1−/−) reduced LV diameters (LVIDs and LVIDd) and increased myocardial wall thickness (LVAWs, LVAWd and LVPWs) with decreased systolic function compared with C57BL/6N controls. e, Alpk3 depletion (Alpk3−/−) increased LV diameters (LVIDs and LVIDd) and decreased systolic function via reduced fractional shortening and ejection fraction, suggesting dilated left ventricle or even dilated cardiomyopathy. The y axis represents the distance (in mm) from the transducer (Vevo 2100); time (in ms) is on the x axis. f, Ap4e1 loss (Ap4e1−/−) caused an impairment of LVIDd and LVIDs and consequently lowered stroke volume. The y axis represents the distance (in mm) from the transducer (Vevo 2100); time (in ms) is on the x axis. g, Representative electrocardiograms from conscious Ap4e1-mutant and C57BL/6N control mice with indication of ECG parameters and interval durations. Ap4e1 loss lowered heart rate and concurrently increased RR interval duration. Data are presented by Mouse Specifics software.