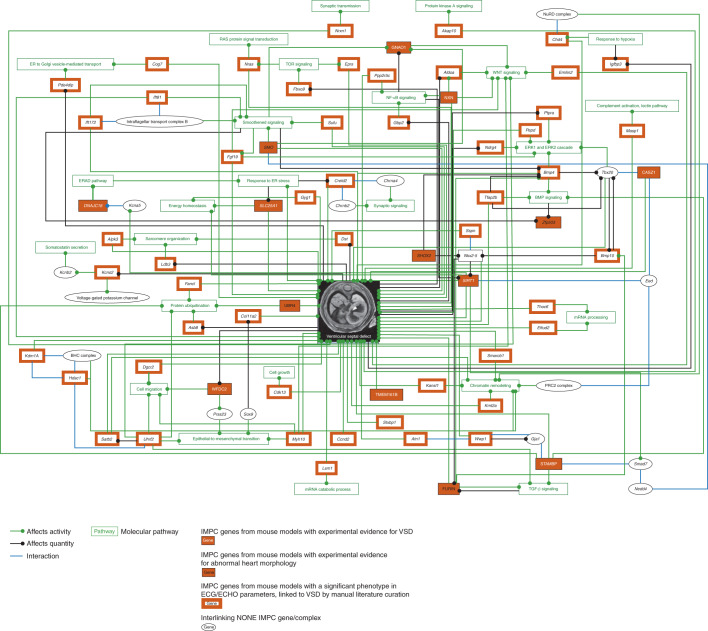
Fig. 2. VSD network.
Ex situ imaging of the embryo heart in homozygous lethal or homozygous subviable single-gene-knockout mice used to identify structural heart defects, such as VSD. Of the mouse lines studied here, 65% were homozygous knockouts (corresponding to LOF in human) and 35% were heterozygous knockouts. This dataset included 248 of 705 lines (35%). Lines were confirmed to be lethal or subviable; we assessed cardiac development by analyzing three-dimensional micro-CT data obtained from iodine contrast-enhanced micro-CT that provides high spatial resolution of up to 3–14 μm per voxel from embryonic day (E)9.5–E18.5 embryos (http://www.mousephenotype.org/data/embryo). No embryo imaging was performed on viable knockout lines. Embryo data were available for only a small subset of the total knockout genes used for this study. The VSD network included nine genes (Sirt1, Stambp, Casz1, Wfdc2, Tmem161b, Nxn, Dnajc18, Gnao1 and Slc25a) that, when LOF was induced, caused early mortality due to structural heart changes but most notably VSDs in the null mutant mice, experimentally shown by computed tomography microscopy. Five more genes (Zpf503 (human ZNF503), Ubr4, Furin, Shox2 and Smo) were associated with cardiac abnormalities after gene depletion, confirmed by gross morphology data. An additional 45 genes were integrated using their association with cardiac malformations and development of a VSD. Only in vivo ECG and TTE data from young adult mice are available because these mutant lines tested positive for viability; therefore, no embryo screening was performed. The network analysis also identified 13 non-IMPC interacting genes. Two transcription factors, NKX2-5 (Furin, Bmp10, Shox2, Sirt1 and Sspn) and TBX20 (Bmp4, Bmp10, Tfap2b and Casz1), known to be important for early cardiac development, were strongly represented. Furthermore, BMP10, a critical regulator of cardiac growth and chamber maturation, chromatin-modifying (Hdac1 and Smarcb1) genes and genes regulated by processes essential for cardiogenesis, for example, smoothened and WNT signaling (Gnao1, Emilin2, Aldoa, Nxn, Smo, Sufu and Ift81), were strongly represented in the network. Compelling evidence of experimental human or rodent data is from relevant publications, primarily peer-reviewed ‘small-scale experiment’ literature used in the network analysis. ECHO, transthoracic echocardiography; ECG, electrocardiography; NONE, genes not analyzed so far in the IMPC; ER, endoplasmic reticulum.