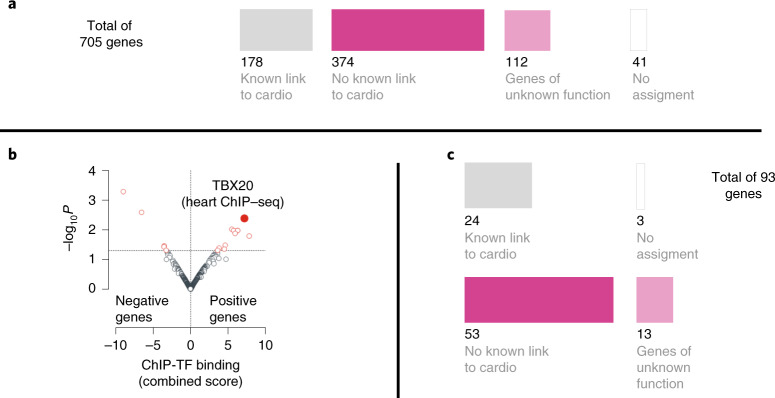
Fig. 3. Alignment and enrichment of 705 IMPC knockout genes.
Using Pharos (https://pharos.nih.gov/), a multimodal web interface, we queried alignment for human cardiac disease relevance. We excluded 41 genes that had no human ortholog and/or limited assignment information from this analysis. This analysis indicated that 155 of the 664 mouse–human orthologous genes were previously linked to heart disease. The remaining 509 orthologous genes had not been previously associated with a cardiac condition and likely represent ‘unappreciated candidate genes’ for CHD. This set of 509 genes was further queried for newness using the Online Mendelian Inheritance in Man (OMIM) catalog (https://omim.org/) and Orphanet, a rare disease portal (https://www.orpha.net/) dataset. This secondary analysis confirmed that 486 of 509 genes were unknown candidate genes and the remaining 23 genes have been associated predominantly with pleiotropic neurodevelopmental disorders that include sporadic congenital heart defects or malformations. a, Alignment of 705 IMPC knockout genes using Pharos, OMIM and Orphanet classified them into genes with a previously known link (178 of 705) or unknown link (486 of 705) to CVD or poorly annotated genes (112 of 486); cardio, cardiovascular diseases and/or heart. b, Genes previously identified with abnormal cardiac function in knockout mice (positive genes) showed strong enrichment (P = 0.004; odds ratio, 1.31; combined score, 7.2) for heart-specific targets of the transcription factor TBX20, a critical regulator of heart development65–67,97, associated with human CHD and adult cardiomyopathies68,70,98,99, while the 3,189 nonsignificant IMPC knockout genes (negative genes) did not. The ChEA 2016 dataset of publicly available ChIP–seq experiments was used for in silico analysis42. Here ‘strong’ enrichment was considered when the following parameters were met: combined score >5 and adjusted P value <0.05; TF, transcription factor. c, Molecular properties of TBX20 target genes showed that 66 of 93 (~70%) have no previous known link to CVD or are poorly annotated genes (13 of 93, ~14%). This observation indicates that, while the role of TBX20 in heart physiology is not completely understood, our dataset sheds light on a previously unknown group of genes with potential relevance for heart development and function.