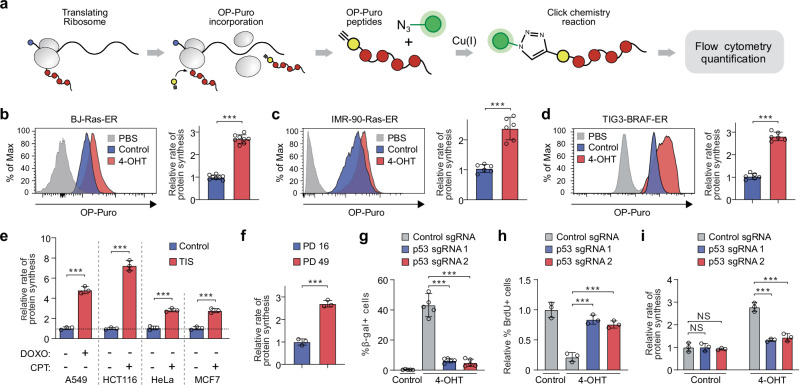
Fig. 1. Global translation rates are elevated in senescent cells.
a Experimental approach to measuring global translation rates in senescent cells. b–d O-propargyl-puromycin (OP-Puro) incorporation and protein synthesis rates in proliferating and senescent BJ-Ras-ER cells (b), IMR-90-Ras-ER cells (c), and TIG3-BRAF-ER cells (d). Data represent mean ± SD from biologically independent experiments (b, n = 8; c, d, n = 6). p-values were calculated using a two-tailed unpaired t test. ***P < 0.001. e Protein synthesis rates in proliferating and senescent cells treated with doxorubicin (DOXO) or camptothecin (CPT) for 7 days. A549, HCT116, HeLa, and MCF7 cells were used. Data represent mean ± SD from biologically independent experiments (n = 3). p-values were calculated using a two-tailed unpaired t test. ***P < 0.001. f OP-Puro incorporation assay in IMR-90 cells undergoing replicative senescence. PD, population doubling. Data represent mean ± SD from biologically independent experiments (n = 3). p-values were calculated using a two-tailed unpaired t test. ***P < 0.001. g Quantification of senescence-associated beta-galactosidase (SA-β-gal) expression in BJ-Ras-ER cells transduced with sgRNAs targeting the p53 gene or a control sequence. Data represent mean ± SD from biologically independent experiments (n = 5). p-values were calculated using a two-tailed unpaired t test. ***P < 0.001. h Quantification of bromodeoxyuridine (BrdU) incorporation in various CRISPR-Cas9-transduced BJ-Ras-ER cells. Data represent mean ± SD from biologically independent experiments (n = 3). p-values were calculated using a two-tailed unpaired t test. ***P < 0.001. i Quantification of protein synthesis rates based on OP-Puro incorporation in proliferating and senescent BJ-Ras-ER cells transduced with either a control sgRNA or sgRNAs targeting the p53 gene. Data represent mean ± SD from biologically independent experiments (n = 3). p-values were calculated using a two-tailed unpaired t test. NS, not significant; ***P < 0.001. Source data, including exact p-values, are provided as Source Data files.